Difference between revisions of "DV RunGroupC Moller"
| Line 75: | Line 75: | ||
| − | == | + | ==[[Alter Phi Angles]]== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
It was shown earlier that the differential cross section scale is <math>\frac{d\sigma}{d\Omega}\approx 16.2\times 10^{-2}mb=16.2\mu b</math> | It was shown earlier that the differential cross section scale is <math>\frac{d\sigma}{d\Omega}\approx 16.2\times 10^{-2}mb=16.2\mu b</math> | ||
Revision as of 00:18, 30 March 2016
need to insert moller shielding into card after moller LUND file is created. (see clas12/beamline)
Simulating the Moller scattering background for EG12
GEANT4 Simulation of Moller Events
Simulation Setup
Determine the Moller background using an LH2 target to check the physics in GEANT4
Distributions For LH2
LH2 Momentum Distribution in the Lab Frame
LH2 Angular Distribution in the Lab Frame
LH2 Momentum Distribution in the Center of Mass Frame
LH2 Angular Distribution in the Center of Mass Frame
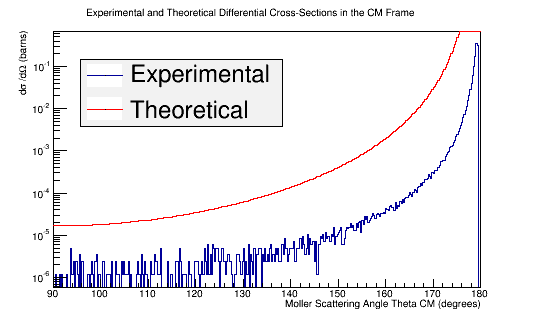
Comparing experimental vs. theoretical for Møller differential cross section 11GeV
Change to a NH3 Target
Replacing the LH2 target with an NH3 target
Distributions for NH3
NH3 Momentum Distribution in the Lab Frame
NH3 Angular Distribution in the Lab Frame
NH3 Momentum Distribution in the Center of Mass Frame
NH3 Angular Distribution in the Center of Mass Frame
LH2 Vs. NH3
Comparing Momentum Distribution in the Lab Frame
Comparing Angular Distribution in the Lab Frame
Comparing Momentum Distribution in the Center of Mass Frame
Comparing Angular Distribution in the Center of Mass Frame
Effects Due to Target Material
Target Density
Atomic Mass and Electron Number Effects
Differential Cross-Section Offset
Reconstruction of Moller Events
Creating LUND Files
Running GEMC
Running Reconstruction Simulations
Analyzing Reconstruction Data
Reconstruction of Random Within Certain Range
Modified gcards
Effects of Solenoid on Moller Electrons
Cover Full Solid Angle of Detector
Calculations of 4-momentum components
Alter Phi Angles
It was shown earlier that the differential cross section scale is
For an Ammonia target:
If the beam had 4E9 incident electrons, the differential cross-section would be found with,
Since extra Phi angles have been produced obviously a larger number of incident electrons would be needed. Looking at the number Moller events are created for 1E6, 1E7, and 4E9 incident electrons, we can estimate the number of incident electrons needed for the number of extra Phi angles produced.
| # of Incident Electrons | # of Moller Events | # of E>500MeV |
|---|---|---|
| 1e6 | 38343 | 134 |
| 1e7 | 383633 | 1490 |
| 4e9 | 12444898 | 48548 |
This shows a trait of providing around 100 Moller electrons of Energy greater that 500 MeV for about 1 million incident electrons of Energy 11 GeV. Since the boosting of the number of Phi angles leaves around 4431573 Moller electrons with Energy greater than 500 MeV, this would imply around 4e10 incident electrons of Energy 11 GeV.
Using the same expression, but this time for 4e10 incident electrons,
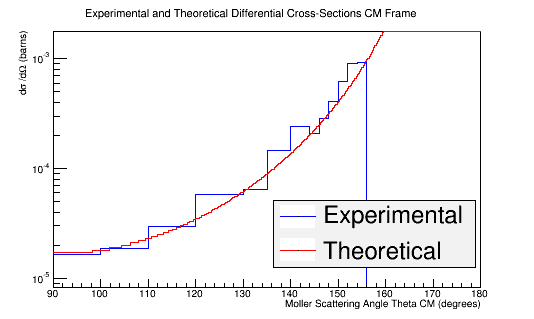
Rebining the histogram to account for the unequal weighting of the bins outlined in the table above
TH1F *Combo=new TH1F("TheoryExperiment","Theoretical and Experimental Differential Cross-Section CM Frame",360,90,180);
Combo->Add(MolThetaCM,8.87e-10);
Combo->Draw();
Double_t Bins[16]={90,100,110,120,130,135,140,142,144,146,148,150,152,154,156,180};
hnew=Combo->Rebin(15,"hnew",Bins);
hnew->Draw();
Theory->Draw("same");
Running LUND files in GEMC
Since the LUND file is limited to 75000 lines, the gemc will have to be run in batch mode;
Creating a batch directory, with two subdirectories; 1)Phi_Parts, 2)submit.
1)Once the LUND file is broken into 178 parts, they can have the LUND extension added by:
prename 's/(Phi_Parts_\d{4})/$1.LUND/' Phi_Parts_*
Placing each of these files into its own directory, within a directory named Phi_Parts
find . -name "*.LUND" -exec sh -c 'mkdir "${1%.*}" ; mv "$1" "${1%.*}" ' _ {} \;
2)Creating the submit directory, and using a c++ program, creating the needed 178
#include <iomanip>
#include <sstream>
#include <iostream>
#include <fstream>
using namespace std;
void submit() {
for(int a=0;a<2;a++)
{
for(int b=0;b<10;b++)
{
for(int c=0;c<10;c++)
{
string filename="submit0";
stringstream hundreds;
hundreds << a;
stringstream tens;
tens << b;
stringstream ones;
ones << c;
string fullname="";
fullname=filename + hundreds.str() + tens.str() + ones.str();
// cout << fullname << "\n";
ofstream myfile;
myfile.open(fullname.c_str());
myfile << "#!/bin/sh\n";
myfile << "#PBS -l nodes=1\n";
myfile << "#PBS -A FIAC\n";
myfile << "#PBS -M vanwdani@isu.edu\n";
myfile << "#PBS -m abe\n";
myfile << "#\n";
myfile << "cd /home/lds/src/CLAS/GEMC\n";
myfile << "tcsh\n";
myfile << "source setup\n";
myfile << "cd /home/lds/src/GEANT/geant4.9.6/geant4.9.6-install/bin/geant4.sh\n";
myfile << "cd /home/vanwdani/src/GEANT4/geant4.9.6/Simulations/Research/Moller/batch/Phi_Parts/Phi_Parts_0";
myfile <<a<<b<<c<<"\n";
myfile << "gemc -USE_GUI=0 -Hall_Material=\"Vacuum\" -INPUT_GEN_FILE=\"LUND, Phi_Parts_0";
myfile <<a<<b<<c;
myfile << ".LUND\" -N=75000 eg12.gcard\n";
myfile << "~/src/CLAS/coatjava-1.0/bin/clas12-reconstruction -i eg12.ev -config DCHB::torus=1.0 ";
myfile << "-config DCHB::solenoid=0.0 -config DCTB::kalman=true -o eg12_rec.ev -s DCHB:DCTB:EC:FTOF:EB\n";
myfile << "~/src/CLAS/coatjava-1.0/bin/rungroovy Analysis.groovy eg12_rec.0.evio\n";
myfile.close();
}
}
}
}
This creates the submitXXXX file
#!/bin/sh #PBS -l nodes=1 #PBS -A FIAC #PBS -M vanwdani@isu.edu #PBS -m abe # cd /home/lds/src/CLAS/GEMC tcsh source setup cd /home/lds/src/GEANT/geant4.9.6/geant4.9.6-install/bin/geant4.sh cd /home/vanwdani/src/GEANT4/geant4.9.6/Simulations/Research/Moller/batch/Phi_Parts/Phi_Parts_0000 gemc -USE_GUI=0 -Hall_Material="Vacuum" -INPUT_GEN_FILE="LUND, Phi_Parts_0000.LUND" -N=75000 eg12.gcard ~/src/CLAS/coatjava-1.0/bin/clas12-reconstruction -i eg12.ev -config DCHB::torus=1.0 -config DCHB::solenoid=0.0 -config DCTB::kalman=true -o eg12_rec.ev -s DCHB:DCTB:EC:FTOF:EB ~/src/CLAS/coatjava-1.0/bin/rungroovy Analysis.groovy eg12_rec.0.evio
Creating a file named lds-submit
Papers used
[1]Farrukh Azfar's Derivation of Moller Scattering
A polarized target for the CLAS detector
An investigation of the spin structure of the proton in deep inelastic scattering of polarized muons on polarized protons
QED Radiative Corrections to Low-Energy Moller and Bhabha Scattering
http://arxiv.org/abs/1602.07609