Difference between revisions of "TF SPIM Intro"
| (23 intermediate revisions by the same user not shown) | |||
| Line 35: | Line 35: | ||
<math>\int_0^{\infty} P(E) dE = \int_0^{\infty} \frac{1}{kT} e^{-\frac{E}{kT}} dE = \frac{1}{kT} \frac{1}{\frac{1}{-kT}} e^{-\frac{E}{kT}} \mid_0^{\infty} = - [e^{-\infty} - e^0]= 1</math> | <math>\int_0^{\infty} P(E) dE = \int_0^{\infty} \frac{1}{kT} e^{-\frac{E}{kT}} dE = \frac{1}{kT} \frac{1}{\frac{1}{-kT}} e^{-\frac{E}{kT}} \mid_0^{\infty} = - [e^{-\infty} - e^0]= 1</math> | ||
| − | <math>P(E=5eV)</math> is calculated by integrating P(E) over some energy interval ( ie:<math> N(v) dv</math>). I will arbitrarily choose 4.9 eV to 5.1 eV as a starting point. | + | Physically, <math>P(E=5eV)</math> is calculated by integrating P(E) over some energy interval ( ie:<math> N(v) dv</math>). I will arbitrarily choose 4.9 eV to 5.1 eV as a starting point. |
| Line 50: | Line 50: | ||
<math>\int_{4.9 eV}^{5.1 eV} P(E) dE = - [e^{-5.1/0.0258} - e^{4.9/0.0258}] = 4.48 \times 10^{-83} - 1.9 \times 10^{-86} \approx 4.48 \times 10^{-83}</math> | <math>\int_{4.9 eV}^{5.1 eV} P(E) dE = - [e^{-5.1/0.0258} - e^{4.9/0.0258}] = 4.48 \times 10^{-83} - 1.9 \times 10^{-86} \approx 4.48 \times 10^{-83}</math> | ||
| − | or in other words | + | or in other words the probability may be approximated by just using the distribution function alone |
<math>P(E=5eV) = e^{-5/0.0258} \approx 10^{-85}</math> | <math>P(E=5eV) = e^{-5/0.0258} \approx 10^{-85}</math> | ||
| Line 56: | Line 56: | ||
This approximation breaks down as <math>E \rightarrow 0.0258 eV</math> | This approximation breaks down as <math>E \rightarrow 0.0258 eV</math> | ||
| − | Since we have 12.011 grams of carbon and 1 mole of carbon = 12.011 g = <math>6 \times 10^{23} </math>carbon atoms | + | Since we have 12.011 grams of carbon and 1 mole of carbon = 12.011 g = <math>6 \times 10^{23} </math>carbon atoms, we would not expect to see a 5 eV carbon atom in a sample size of <math>6 \times 10^{23} </math> carbon atoms when the probability of observing such an atom is <math>\approx 10^{-85}</math>. Note: The mass of the earth is about <math>10^{27}</math> g <math>\approx 10^{50}</math> atoms, so a carbon atom with an energy of 5 eV would be difficult to observe in a detector the size of the earth . |
| − | + | The average energy we expect to see would be calculated by | |
| − | |||
| − | The energy we expect to see would be calculated by | ||
<math><E> = \int_{0}^{\infty} E \cdot P(E) dE</math> | <math><E> = \int_{0}^{\infty} E \cdot P(E) dE</math> | ||
| Line 105: | Line 103: | ||
The above particle detector was an example of describing a stochastic process using a probability distribution to determine the likely hood of finding an atom with a certain energy. | The above particle detector was an example of describing a stochastic process using a probability distribution to determine the likely hood of finding an atom with a certain energy. | ||
| − | + | Physics at the Quantum Mechanics scale contains some of the clearest examples of such a non-deterministic systems. The canonical systems in Thermodynamics is another example. | |
Basically the monte-carlo method uses a random number generator (RNG) to generate a distribution (gaussian, uniform, Poission,...) which is used to solve a stochastic process based on an astochastic description. | Basically the monte-carlo method uses a random number generator (RNG) to generate a distribution (gaussian, uniform, Poission,...) which is used to solve a stochastic process based on an astochastic description. | ||
| Line 170: | Line 168: | ||
Step | Step | ||
| − | # login to | + | # login to minerve.<br> [http://physics.isu.edu/~tforest/Classes/NucSim/Day1/XwindowsOnWindows.html click here for a description of logging in if using windows] otherwise (ssh username@minerve.cose.isu.edu) |
# mkdir src | # mkdir src | ||
# cd src | # cd src | ||
| − | # | + | # scp -r foretony@minerve.cose.isu.edu:src/PI ./ |
# ls | # ls | ||
| − | # cd | + | # cd PI |
# make | # make | ||
| − | #./ | + | #./PI_MC |
| − | [http://physics.isu.edu/~tforest/Classes/NucSim/Day1/RNG/Marsaglia/noviceExample/ Here is a web link to the source files you can copy in case the above doesn't work] | + | [http://physics.isu.edu/~tforest/Classes/NucSim/Day1/RNG/Marsaglia/noviceExample/ Here is a web link to the source files you can copy in case the above doesn't work] in this case the final command is "./rnd_test" |
== A Root Primer == | == A Root Primer == | ||
| Line 193: | Line 191: | ||
$ROOTSYS/bin/root | $ROOTSYS/bin/root | ||
| + | |||
| + | another method | ||
| + | |||
| + | source ~foretony/src/ROOT/root/bin/thisroot.sh | ||
| + | |||
=== Example 1: Create Ntuple and Draw Histogram=== | === Example 1: Create Ntuple and Draw Histogram=== | ||
| Line 199: | Line 202: | ||
=== Definitions === | === Definitions === | ||
;Total cross section | ;Total cross section | ||
| − | :<math>\sigma \equiv \frac{\# particles | + | :<math>\sigma \equiv \frac{\# \mbox{ particles scattered}} {\frac{ \# \mbox{ incident particles}}{\mbox{Area}}}</math> |
;Differential cross section | ;Differential cross section | ||
| − | :<math>\sigma(\theta)</math> = <math> \frac{d \sigma}{d \Omega} \equiv \frac{\frac{\# particles | + | :<math>\sigma(\theta)</math> = <math> \frac{d \sigma}{d \Omega} \equiv \frac{\frac{\# \mbox{ particles scattered}}{\mbox{solid angle}}} {\frac{ \# \mbox{incident particles}}{\mbox{Area}}}</math> |
; Solid Angle | ; Solid Angle | ||
| Line 210: | Line 213: | ||
:A= surface area of detector | :A= surface area of detector | ||
:r=distance from interaction point to detector | :r=distance from interaction point to detector | ||
| − | :<math>\Omega = \frac{A}{r^2} </math> | + | :<math>\Omega = \frac{A}{r^2} sr </math> |
| − | : <math>A_{sphere} = 4 \pi r^2</math> if your detector was a hollow ball | + | :<math>sr \equiv</math> steradians |
| − | :<math>\Omega_{max} = \frac{4 \pi r^2}{r^2} = 4\pi</math> | + | : <math>A_{\mbox{sphere}} = 4 \pi r^2</math> if your detector was a hollow ball |
| + | :<math>\Omega_{\mbox{max}} = \frac{4 \pi r^2}{r^2} = 4\pi</math>steradians | ||
;Units | ;Units | ||
:Cross-sections have the units of Area | :Cross-sections have the units of Area | ||
:1 barn = <math>10^{-28} m^2</math> | :1 barn = <math>10^{-28} m^2</math> | ||
| − | ; [units of <math>\sigma(\theta)</math>] =<math>\frac{\frac{[particles]}{[ | + | ; [units of <math>\sigma(\theta)</math>] =<math>\frac{\frac{[\mbox{particles}]}{[\mbox{steradian}]}} {\frac{ [ \mbox{particles}]}{[m^2]}} = m^2</math> |
;Luminosity | ;Luminosity | ||
| − | :<math>L = \frac{Number of Scatterers}{Area \cdot time} \sim i_{beam} \rho_{target} l_{target}</math> | + | :<math>L = \frac{\mbox{Number of Scatterers}}{\mbox{Area} \cdot \mbox{time}} \sim i_{\mbox{beam}} \rho_{\mbox{target}} l_{\mbox{target}}</math> |
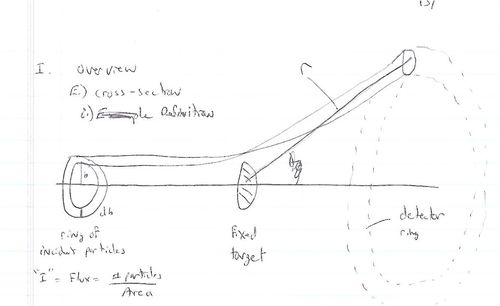
| − | [[Image:FixedTargetScatteringCrossSection.jpg | | + | [[Image:FixedTargetScatteringCrossSection.jpg | 500 px]] |
; Fixed target scattering | ; Fixed target scattering | ||
: <math>N_{in}</math>= # of particles in = <math>I \cdot A_{in}</math> | : <math>N_{in}</math>= # of particles in = <math>I \cdot A_{in}</math> | ||
| Line 236: | Line 240: | ||
;Units | ;Units | ||
| − | : <math>[d N] = [\frac {particles}{m^2}][m^2] [ | + | : <math>[d N] = [\frac {\mbox{particles}}{m^2}][m^2] [\mbox{steradian}] </math> = # of particles |
: or for a count rate divide both sides by time and you get beam current on the RHS | : or for a count rate divide both sides by time and you get beam current on the RHS | ||
: integrate and you have the total number of counts | : integrate and you have the total number of counts | ||
| Line 253: | Line 257: | ||
This example is an example of classical scattering. | This example is an example of classical scattering. | ||
| − | Our goal is to find <math>\sigma(\theta)</math> for an elastic collision of 2 impenetrable spheres of diameter <math>a</math>. We need to look for a relationship between the impact parameter<math> b</math> and the scattering angle<math> \theta</math>. To find this relationship, let's solve this elastic scattering problem by describing the collision using the Center of Mass (C.M.) coordinate system in terms of the reduced mass. As we shall see, | + | Our goal is to find <math>\sigma(\theta)</math> for an elastic collision of 2 impenetrable spheres of diameter <math>a</math>. We need to look for a relationship between the impact parameter <math> b</math> and the scattering angle <math> \theta</math>. To find this relationship, let's solve this elastic scattering problem by describing the collision using the Center of Mass (C.M.) coordinate system in terms of the reduced mass. As we shall see, the 2-body collision becomes a 1-body problem when a C.M. coordinate system is used. Then we will describe the motion of the reduced mass in the C.M. Frame. |
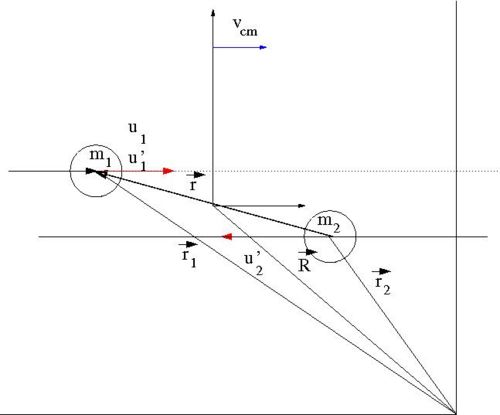
[[Image:SPIM_ElasCollis_Lab_CM_Frame.jpg | 500 px]] | [[Image:SPIM_ElasCollis_Lab_CM_Frame.jpg | 500 px]] | ||
| Line 295: | Line 299: | ||
: <math>\vec{r}_1 = \frac{\mu}{m_1} \vec{r}</math> | : <math>\vec{r}_1 = \frac{\mu}{m_1} \vec{r}</math> | ||
| − | : <math>\vec{r}_2 = \frac{\mu}{m_2} \vec{r}</math> | + | : <math>\vec{r}_2 = -\frac{\mu}{m_2} \vec{r}</math> |
We can use the above reduced mass relationships to construct the Lagrangian in terms of <math>\vec{r}</math> instead of <math>\vec{r}_1</math> and <math> \vec{r}_2</math> thereby reducing the problem from a 2-body problem to a 1-body problem. | We can use the above reduced mass relationships to construct the Lagrangian in terms of <math>\vec{r}</math> instead of <math>\vec{r}_1</math> and <math> \vec{r}_2</math> thereby reducing the problem from a 2-body problem to a 1-body problem. | ||
| Line 368: | Line 372: | ||
:or | :or | ||
: <math>d \phi = \frac{\ell}{\mu r^2 \dot{r}} dr</math> | : <math>d \phi = \frac{\ell}{\mu r^2 \dot{r}} dr</math> | ||
| + | |||
| + | |||
We now need an expression for <math>\dot{r}</math> in order to integrate the above equation to determine the functional dependence of <math>\phi</math> and hence<math> \theta</math>. | We now need an expression for <math>\dot{r}</math> in order to integrate the above equation to determine the functional dependence of <math>\phi</math> and hence<math> \theta</math>. | ||
| + | |||
| + | The potential in the Lagrangian though is infinite for <math>r \le a</math> . Let's use the property of conservation of energy to accommodate this mathematical construct. | ||
Since Energy is conserved (Elastic Scattering), we may define the Hamiltonian as | Since Energy is conserved (Elastic Scattering), we may define the Hamiltonian as | ||
| − | <math>H = T + U = \frac{1}{2} (mu \dot{r}^2 + \frac{\ell}{\mu r^2}) + U(r) = constant \equiv E</math> | + | <math>H = T + U = \frac{1}{2} (\mu \dot{r}^2 + \frac{\ell}{\mu r^2}) + U(r) = constant \equiv E</math> |
solving for <math>\dot{r}</math> | solving for <math>\dot{r}</math> | ||
Latest revision as of 14:54, 2 September 2016
Introduction
Experimentalists use simulations to predict the sources of background which will interfere with the signal they plan on measuring. An important aspect of this process is to understand how signals are produced in your measurement device. Devices share the common problem of isolating a signal produced in the device from the noise that is present in the device.
Below is a description of how signals are produced in bulk materials.
Particle Detection
A device detects a particle only after the particle transfers energy to the device.
Energy intrinsic to a device depends on the material used in a device
Consider a device made of some material with an average atomic number () at some temperature (). The material's atoms are in constant thermal motion (unless you can manage to have T = zero degrees Klevin).
Statistical Thermodynamics tells us that the canonical energy distribution of the atoms is given by the Maxwell-Boltzmann statistics such that
represents the probability of any atom in the system having an energy where
Note: You may be more familiar with the Maxwell-Boltzmann distribution in the form
where would represent the molecules in the gas sample with speeds between and
Example 1: P(E=5 eV)
- What is the probability that an atom in a 12.011 gram block of carbon would have an energy of 5 eV?
First lets check that the probability distribution is Normalized; ie: does ?
Physically, is calculated by integrating P(E) over some energy interval ( ie:). I will arbitrarily choose 4.9 eV to 5.1 eV as a starting point.
assuming a room temperature of
then
and
or in other words the probability may be approximated by just using the distribution function alone
This approximation breaks down as
Since we have 12.011 grams of carbon and 1 mole of carbon = 12.011 g = carbon atoms, we would not expect to see a 5 eV carbon atom in a sample size of carbon atoms when the probability of observing such an atom is . Note: The mass of the earth is about g atoms, so a carbon atom with an energy of 5 eV would be difficult to observe in a detector the size of the earth .
The average energy we expect to see would be calculated by
If you used this block of carbon as a detector you would easily notice an event in which a carbon atom absorbed 5 eV of energy as compared to the energy of a typical atom in the carbon block.
- Silicon detectors and Ionization chambers are two commonly used devices for detecting radiation.
approximately 1 eV of energy is all that you need to create an electron-ion pair in Silicon
approximately 10 eV of energy is needed to ionize an atom in a gas chamber
The low probability of having an atom with 10 eV of energy means that an ionization chamber would have a better Signal to Noise ratio (SNR) for detecting 10 eV radiation than a silicon detector
But if you cool the silicon detector to 200 degrees Kelvin (200 K) then
So cooling your detector will slow the atoms down making it more noticable when one of the atoms absorbs energy.
also, if the radiation flux is large, more electron-hole pairs are created and you get a more noticeable signal.
Unfortunately, with some detectore, like silicon, you can cause radiation damage that diminishes it's quantum efficiency for absorbing energy.
- What does this have to do with Simulations?
- You just did a SImulation. Consider the following description of the Monte Carlo Method
The Monte Carlo method
- Stochastic
- from the greek word "stachos"
- a means of, relating to, or characterized by conjecture and randomness.
A stochastic process is one whose behavior is non-deterministic in that the next state of the process is partially determined.
The above particle detector was an example of describing a stochastic process using a probability distribution to determine the likely hood of finding an atom with a certain energy.
Physics at the Quantum Mechanics scale contains some of the clearest examples of such a non-deterministic systems. The canonical systems in Thermodynamics is another example.
Basically the monte-carlo method uses a random number generator (RNG) to generate a distribution (gaussian, uniform, Poission,...) which is used to solve a stochastic process based on an astochastic description.
Example 2 Calculation of
- Astochastic description
- may be measured as the ratio of the area of a circle of radius divided by the area of a square of length
You can measure the value of if you physically measure the above ratios.
- Stochastic description
- Construct a dart board representing the above geometry, throw several darts at it, and look at a ratio of the number of darts in the circle to the total number of darts thrown (assuming you always hit the dart board).
- Monte-Carlo Method
- Here is an outline of a program to calulate using the Monte-Carlo method with the above Stochastic description
begin loop x=rnd y=rnd dist=sqrt(x*x+y*y) if dist <= 1.0 then numbCircHits+=1.0 numbSquareHist += 1.0 end loop print PI = 4*numbCircHits/numbSquareHits
A Unix Primer
To get our feet wet using the UNIX operating system, we will try to solve example 2 above using a RNG under UNIX
List of important Commands
- ls
- pwd
- cd
- df
- ssh
- scp
- mkdir
- printenv
- emacs, vi, vim
- make, gcc
- man
- less
- rm
Most of the commands executed within a shell under UNIX have command line arguments (switches) which tell the command to print information about using the command to the screen. The common forms of these switches are "-h", "--h", or "--help"
ls --help ssh -h
the switch deponds on your flavor of UNIX
if using the switch doesn't help you can try the "man" (sort for manual) pages (if they were installed). Try
man -k pwd
the above command will search the manual for the key word "pwd"
Example 3: using UNIX to compile a RNG
Step
- login to minerve.
click here for a description of logging in if using windows otherwise (ssh username@minerve.cose.isu.edu) - mkdir src
- cd src
- scp -r foretony@minerve.cose.isu.edu:src/PI ./
- ls
- cd PI
- make
- ./PI_MC
Here is a web link to the source files you can copy in case the above doesn't work in this case the final command is "./rnd_test"
A Root Primer
In bash shell do
export ROOTSYS=~tforest/src/ROOT/root
if chsh do
setenv ROOTSYS ~tforest/src/ROOT/root
To start the root program type
$ROOTSYS/bin/root
another method
source ~foretony/src/ROOT/root/bin/thisroot.sh
Example 1: Create Ntuple and Draw Histogram
Cross Sections
Definitions
- Total cross section
- Differential cross section
- =
- Solid Angle
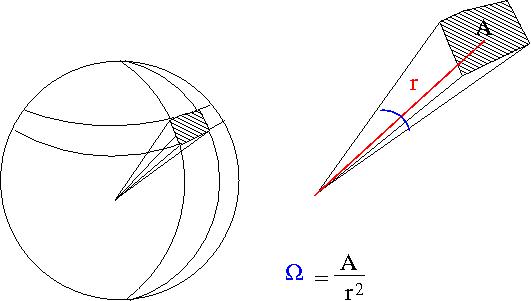
- = surface area of a sphere covered by the detector
- ie;the detectors area projected onto the surface of a sphere
- A= surface area of detector
- r=distance from interaction point to detector
- steradians
- if your detector was a hollow ball
- steradians
- Units
- Cross-sections have the units of Area
- 1 barn =
- [units of ] =
- Luminosity
- Fixed target scattering
- = # of particles in =
- is the area of the ring of incident particles
- = # particles in a ring of radius and thickness
You can measure if you measure the # of particles detected in a known detector solid angle from a known incident particle Flux () as
Alternatively if you have a theory which tells you which you want to test experimentally with a beam of flux then you would measure counts (particles)
- Units
- = # of particles
- or for a count rate divide both sides by time and you get beam current on the RHS
- integrate and you have the total number of counts
- Classical Scattering
- In classical scattering you get the same number of particles out that you put in (no capture, conversion,..)
- tells you how the impact parameter changes with scattering angle
Example 4: Elastic Scattering
This example is an example of classical scattering.
Our goal is to find for an elastic collision of 2 impenetrable spheres of diameter . We need to look for a relationship between the impact parameter and the scattering angle . To find this relationship, let's solve this elastic scattering problem by describing the collision using the Center of Mass (C.M.) coordinate system in terms of the reduced mass. As we shall see, the 2-body collision becomes a 1-body problem when a C.M. coordinate system is used. Then we will describe the motion of the reduced mass in the C.M. Frame.
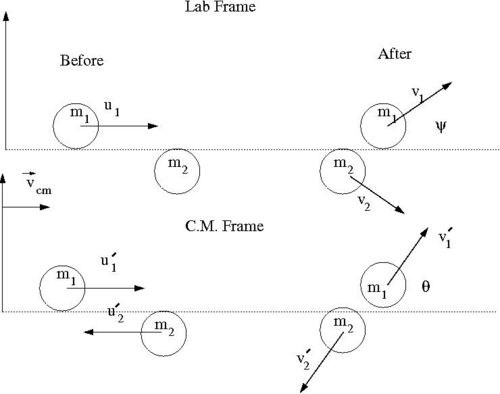 Media:SPIM_ElasCollis_Lab_CM_Frame.xfig.txt
Media:SPIM_ElasCollis_Lab_CM_Frame.xfig.txt
- Variable definitions
- = impact parameter ; distance of closest approach
- = mass of incoming ball
- = mass of target ball
- = iniital velocity of incoming ball in Lab Frame
- = final velocity of in Lab Frame
- = scattering angle of in Lab frame after collision
- = iniital velocity of in C.M. Frame
- = final velocity of in C.M. Frame
- = iniital velocity of in C.M. Frame
- = final velocity of in C.M. Frame
- = scattering angle of in C.M. frame after collision
- Determining the reduced mass
- vector definitions
- = a position vector pointing to the location of
- = a position vector pointing to the location of
- = a position vector pointing to the center of mass of the two ball system
- = the magnitude of this vector is the distance between the two masses
In the C.M. reference frame the above vectors have the following relationships
solving the above equations for and and defining the reduced mass as
- reduced mass
leads to
We can use the above reduced mass relationships to construct the Lagrangian in terms of instead of and thereby reducing the problem from a 2-body problem to a 1-body problem.
- Construct the Lagrangian
The Lagrangian is defined as:
where
kinetic energy of the system
Potential energy of the system which describes the interaction
- =
- =
after substituting derivative of the expressions for and
- =
The 2-body problem is now described by a 1-body Lagrangian we need to determine which coordinate system (cartesian, spherical,..) to use to write an expression for (). Polar seems best unless there is a dependence in the azimuthal angle.
Lagranges equations of motion are given by
where represents one of the coordinate (cannonical variables).
To get the classical scattering cross section we are interested in finding an expression for the dependence of the impact parameter on the scattering angle,.
Now lets redraw the collision in terms of a reference frame fixed on (before collision its the Lab Frame but not after collision).
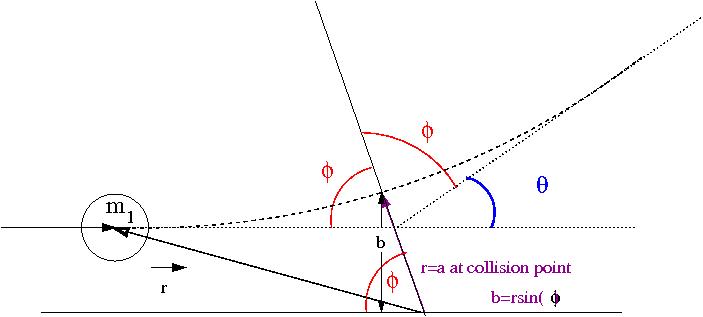 Media:SPIM_ElasColls_CMFrame_xfig.txt
Media:SPIM_ElasColls_CMFrame_xfig.txt
The C.M. Frame rides along the center of mass, the above coordinate system though has its origin on . The above drawing identifies and for the system at the point of the collision in which the CM frame is a distance (the size of the ball) from the origin of the coordinate system fixed to . If then there is no collision (), otherwise a collision happens when r=a (the distance between the balls is equal to their diameter). A head on collision is defined as ().
- Observation
- as gets smaller, gets bigger
Using plane polar coordinates () we can describe the problem in the lab frame as:
Lagranges Equation of Motion:
there is a constant of motion ( Constant angular momentum)
substitute into
The two equations above are in terms of and whereas our goal is to find an expression for . Since is related to and is related to (; see figure above) we should try and find expressions for in terms of
- Trick
- or
We now need an expression for in order to integrate the above equation to determine the functional dependence of and hence.
The potential in the Lagrangian though is infinite for . Let's use the property of conservation of energy to accommodate this mathematical construct.
Since Energy is conserved (Elastic Scattering), we may define the Hamiltonian as
solving for
substituting the above into the equation for and integrating:
For :
substituting this expression for into the last expression for above :
- Integral Table
let
then
or
- Now substitute the above into the expression for
drop the negative sign, sqrt in denominator allows this, and use the trig identity
- compare with result from definition
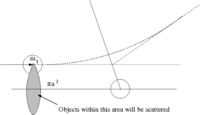
- = scattering cross-section
- number of particles scattered = number of incident particles
- Area = = The area profile in which a collision occurs( the ball diameter is )
Lab Frame Cross Sections
The C.M. frame is often chosen to theoretically calculate cross-sections even though experiments are conducted in the Lab frame. In such cases you will need to transform cross-sections between two frames.
The total cross-section should be frame independent
or
where
is in the CM frame and is in the Lab frame.
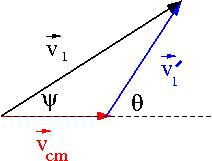
- A non-relativistic transformation
The transformation is governed by the dependence of on
Lets return back to our picture of the scattering Process
if we superimpose the vectors and we have
Trig identities (non-relativistic Gallilean transformation) tell us
solving for
For an elastic collision only the directions change in the CM Frame: &
- From the definition of the C.M.
- conservation of momentum in CM Frame
- Gallilean Coordinate transformation
- another expression for
using the above gallilean transformation we can do the following
or
after a little trig substitution
constant
now use the chain rule to find
- constant
after substitution:
For the above equation to be more useful one would prefer to recast it in terms of only and masses.