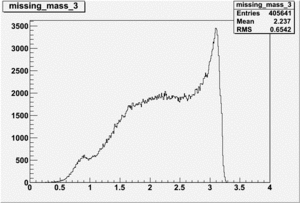
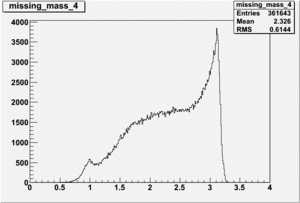
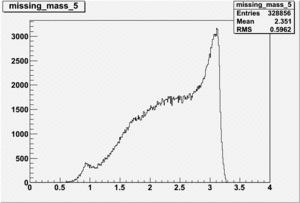
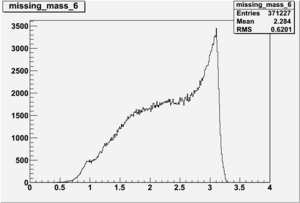
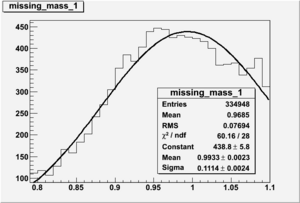
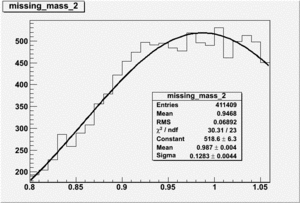
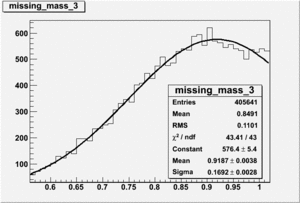
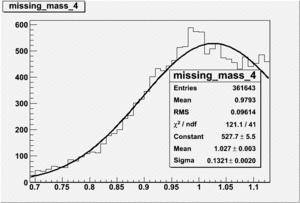
DeltaDoverD Progress
9/5/08
SIDIS Analysis
a.) Cross-Section comparison
i.) Calculate absolute cross section for = 0.5, 1 < Q^2 < 4 GeV^2 , W = 1.45 +/- 0.2 GeV
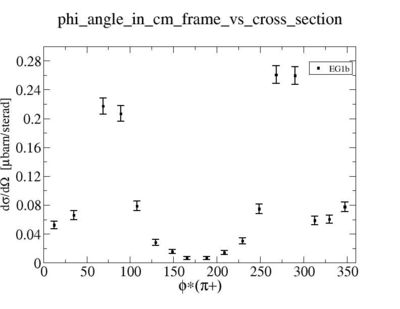
ii.) Plot -vs- when = -0.3 also plot -vs-
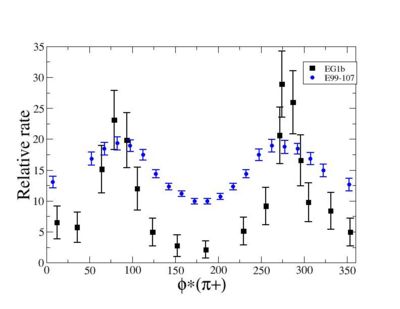
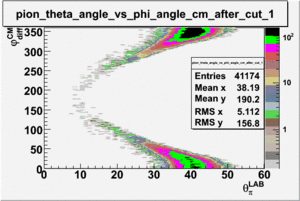
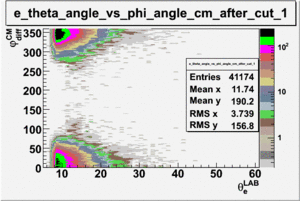
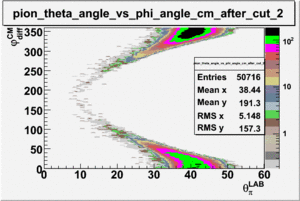
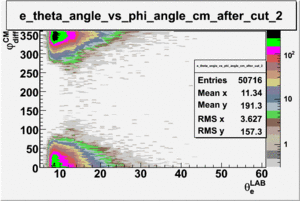
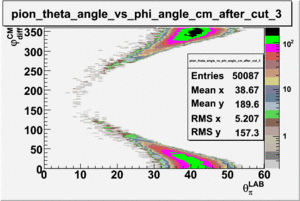
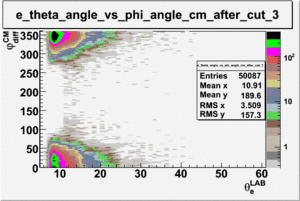
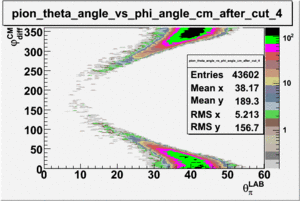
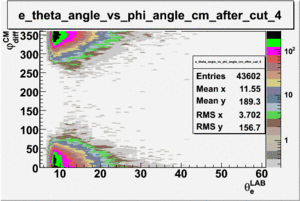
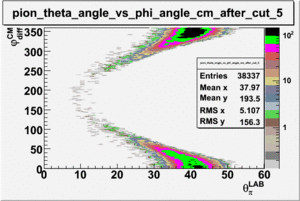
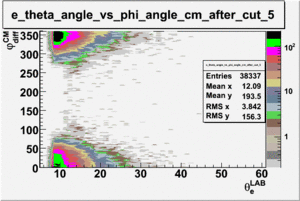
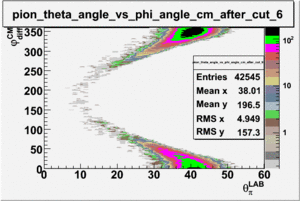
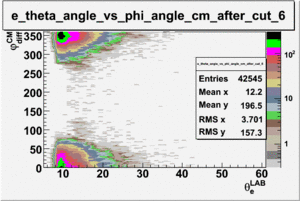
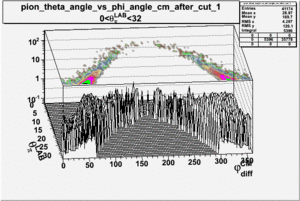
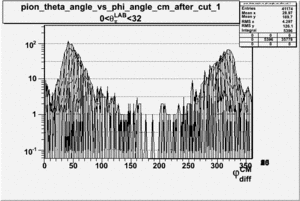
As one can see from histograms of the pion_theta_angle_vs_phi_angle_in_CM_Frame and the electron_theta_angle_vs_phi_angle_in_CM_Frame, the pion and electron acceptance in the region of is nearly zero(significantly low).
_vs_ plot shows that the cut should not make much difference on plot. The cut around should reduce the number of pions around 0 and 360 of phi angle.
9/19/08
SIDIS Analysis
a.) Cross-Section comparison
i.) Calculate absolute cross section for = 0.5, 1 < Q^2 < 4 GeV^2 , W = 1.45 +/- 0.2 GeV
?
- Documentation
The five-fold differential cross section for single pion production is equal to the following:
The Jacobian term can be given as:
where is the virtual photon flux and can be written as
The reason of having low cross section might be binning
In paper they use the following number of bins:
| Variable | Num. Bin | Range | Bin Size |
| W | 27 | 1.15 - 1.7 GeV | 20 MeV |
| 7 | 1.1 - 5.0 | variable | |
| 10 | -1.0 - 1.0 | 0.2 | |
| 24 | -180. - 180 | 15 |
Number of bins in my case were 360.
Number of bins equal is ~24
When i applied cut, the number of entries were reduced to 152.
In paper they use liquid-hydrogen unpolarized target(we have NH3 and ND3 polarized). The polarized electron beam current is 8 nA(in our case it is about 6 nA). and luminosity is different too. It also depends on statistics.
ii.) What is smallest angular coverage of EC (8 degrees?) Determine phi region where there is no acceptance ( where should we stop plotting data)?
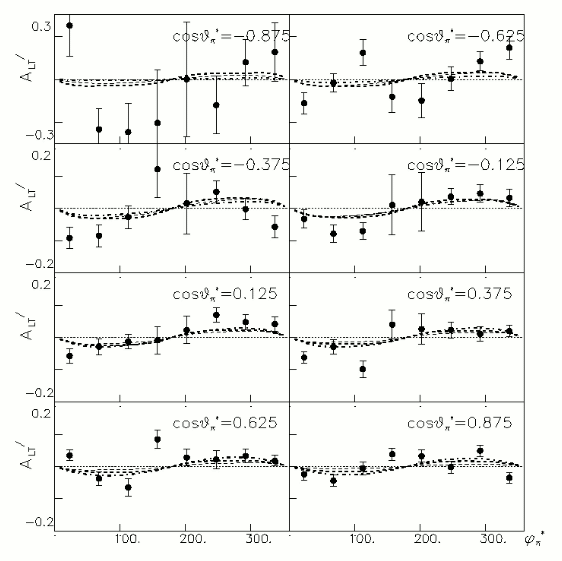
iii.) Asymmetry Calculation
a.) Beam Asymmetry Plot
In order to plot Beam Asymmetry we need to plot for the first time the histogram of the invariant_mass_vs_cross_section. Then determine the structure function fitting the cross section and calculating the beam asymmetry function which is given as :
for given cos(theta) of the pion in cm, Q^2 and W invariant mass.
Choose kinematics ( a single theta and phi point )to max our stats and comparison to paper Histogram the following
- Number of e-pi coincidence events, number of FC counts
- Number of e-pi coincidence events/FC counts
- Number of e-pi coincidence events/FC count/Pt
for groups of runs with h_e,P_t = ++, +-, -+, --
table with 4 columns of h_e, P_t and 3 rows of the above histograms
9/26/08
SIDIS Analysis
1.) make semi-inclusive spectrum:
a.) h>0 Pt>0 b.) h > 0 pt<0 c.)h<0 pt>0 d.)h<0 pt<0
2.) ad up opposite target polarization histograms
3.) subtract h> 0 and h<0
helflag = 1 =>
helflag = 2 =>
helflag = 3 =>
helflag = 4 =>
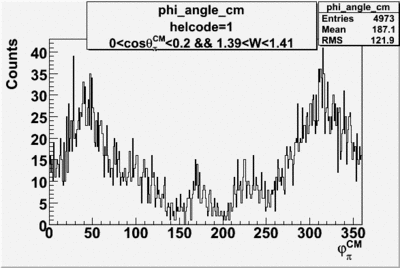
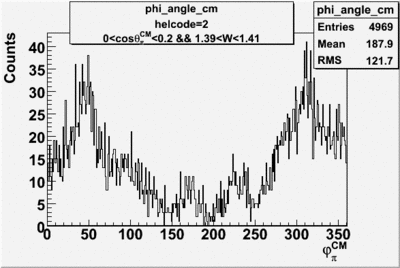
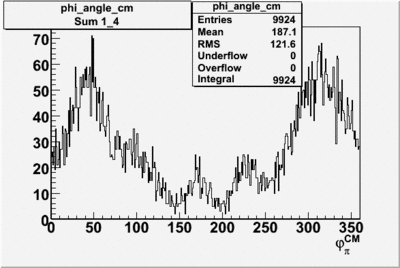
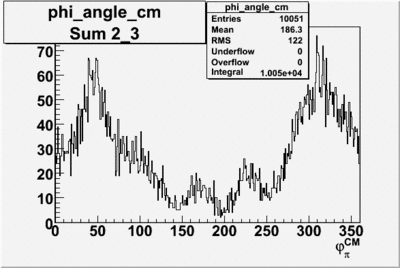
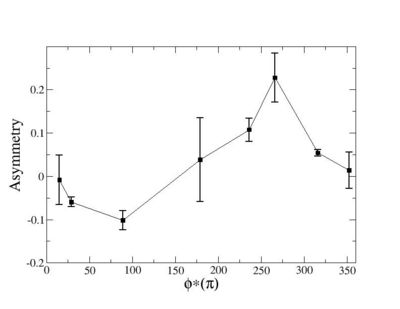
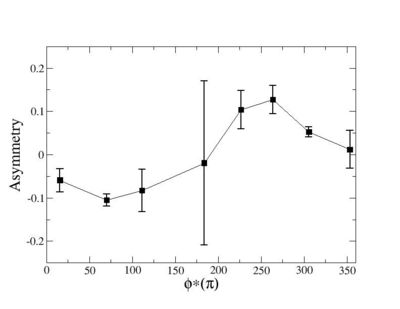
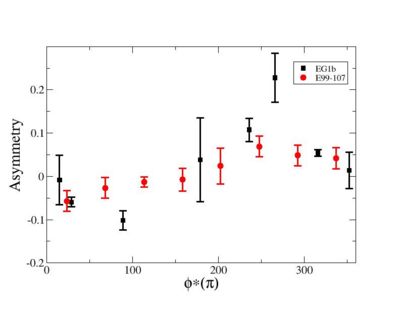
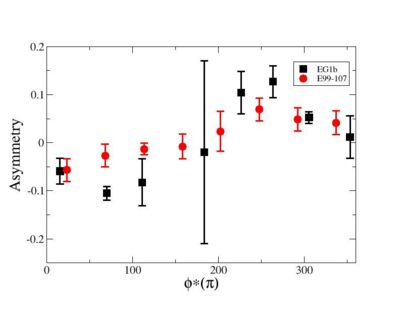
Phi_angle_CM vs Asymmetry
Compared data(there is no table for asymmetry values like cross section):
11/21/08
0.) insert run summarry table, Pb, Pt, PB*Pt, Btorus
1.) make semi-inclusive spectrum:
a.) h>0 Pt>0
b.) h > 0 pt<0
c.)h<0 pt>0
d.)h<0 pt<0
2.) helicity difference plots for Pt>0 and Pt<0
3.) Asym plots for Pt>0 and Pt< 0
4.) unpolarized target asymmetry
Rebin asymmetry hisograms and combine the two into a total asymmetry histogram
Run Summary Table
The table below should have run ranges with according to the elastic asymmetry measurements stored at http://www.jlab.org/Hall-B/secure/eg1/EG2000/eg1b_analysis_progress.htm.
http://www.jlab.org/Hall-B/secure/eg1/EG2000/josh/pbpt/
5.73 in
pos = 0.45743 +/- 0.04269
neg = -0.38816 +/- 0.04556
5.73 out
pos = 0.46604 +/- 0.03496
neg = -0.50684 +/- 0.03578
| Start Run Number | Beam Polarization(Pb) | Target Polarization(Pt) from dstdump file | Pb*Pt | Beam Torus | Beam Energy (MeV) |
| 26998 | 0 | -0.69 | 0.46604 | -2250 | 5736 |
| 26996 | -0.71 | -0.69 | 0.46604 | -2250 | 5736 |
| 26995 | -0.71 | -0.69 | 0.46604 | -2250 | 5736 |
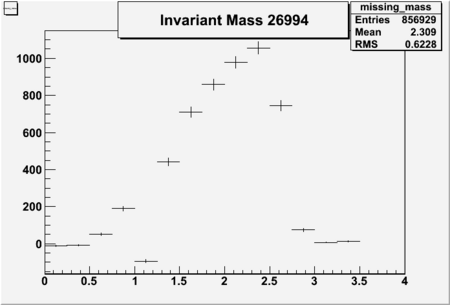
| 26994 | -0.71 | -0.73 | 0.46604 | -2250 | 5736 |
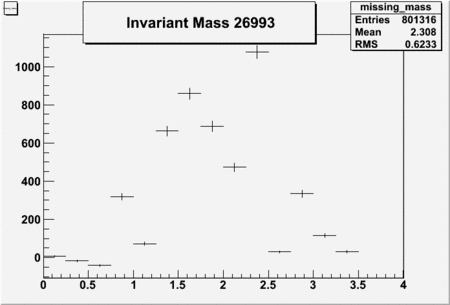
| 26993 | -0.71 | -0.72 | 0.46604 | -2250 | 5736 |
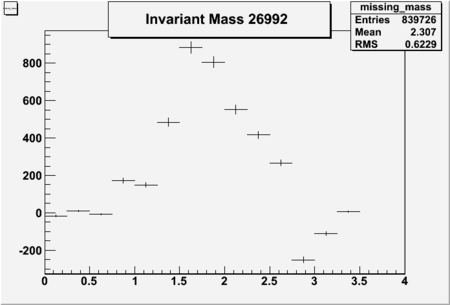
| 26992 | -0.71 | -0.73 | 0.46604 | -2250 | 5736 |
| 26991 | -0.71 | -0.75 | 0.46604 | -2250 | 5736 |
| 26990 | -0.71 | -0.76 | 0.46604 | -2250 | 5736 |
| 26989 | 0.71 | -0.67 | -0.38816 | 2250 | 5736 |
| 26988 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26987 | 0.71 | -0.67 | -0.38816 | 2250 | 5736 |
| 26986 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26985 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26984 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26983 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26981 | 0.71 | -0.68 | -0.38816 | 2250 | 5736 |
| 26980 | 0.71 | -0.7 | -0.38816 | 2250 | 5736 |
| 26979 | 0.71 | -0.71 | -0.38816 | 2250 | 5736 |
| 26966 | 0.71 | 0.74 | 0.45743 | 2250 | 5736 |
| 26965 | 0.71 | 0.74 | 0.45743 | 2250 | 5736 |
| 26964 | 0.71 | 0.74 | 0.45743 | 2250 | 5736 |
| 26963 | 0.71 | 0.74 | 0.45743 | 2250 | 5736 |
| 26962 | 0.71 | 0.74 | 0.45743 | 2250 | 5736 |
| 26961 | 0.71 | 0.75 | 0.45743 | 2250 | 5736 |
| 26960 | 0.71 | 0.76 | 0.45743 | 2250 | 5736 |
| 26959 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26958 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26957 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26956 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26955 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26954 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26953 | 0.71 | 0.69 | 0.45743 | 2250 | 5736 |
| 26952 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26951 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26948 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26947 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26946 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26945 | 0.71 | 0.7 | 0.45743 | 2250 | 5736 |
| 26943 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26942 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26941 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26940 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26939 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26938 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26937 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26934 | 0.7 | 0.7 | 0.45743 | 2250 | 5736 |
| 26933 | 0.7 | 0.71 | 0.45743 | 2250 | 5736 |
| 26932 | 0.7 | 0.71 | 0.45743 | 2250 | 5736 |
| 26931 | 0.7 | 0.71 | 0.45743 | 2250 | 5736 |
| 26930 | 0.7 | 0.71 | 0.45743 | 2250 | 5736 |
| 26929 | 0.7 | 0.71 | 0.45743 | 2250 | 5736 |
| 26928 | 0.7 | 0.72 | 0.45743 | 2250 | 5736 |
| 26927 | 0.7 | 0.72 | 0.45743 | 2250 | 5736 |
| 26926 | 0.7 | 0.73 | 0.45743 | 2250 | 5736 |
| 26925 | 0.7 | 0.73 | 0.45743 | 2250 | 5736 |
| 27074 | 0.71 | -0.81 | -0.38816 | 2250 | 5736 |
| 27075 | 0.71 | -0.77 | -0.38816 | 2250 | 5736 |
| 27076 | 0.71 | -0.74 | -0.38816 | 2250 | 5736 |
| 27077 | 0.71 | -0.74 | -0.38816 | 2250 | 5736 |
| 27078 | 071 | -0.75 | -0.38816 | 2250 | 5736 |
| 27079 | 0.71 | -0.72 | -0.38816 | 2250 | 5736 |
| 27100 | 0.71 | 0.76 | 0.45743 | 2250 | 5736 |
| 27101 | 0.71 | 0.76 | 0.45743 | 2250 | 5736 |
| 27102 | 0.71 | 0.73 | 0.45743 | 2250 | 5736 |
| 27105 | 0.61 | 0.73 | 0.45743 | 2250 | 5736 |
| 27106 | 0.61 | 0.73 | 0.45743 | 2250 | 5736 |
| 27107 | 0.61 | -0.71 | -0.38816 | 2250 | 5736 |
| 27108 | 0.61 | -0.7 | -0.38816 | 2250 | 5736 |
| 27109 | 0.61 | -0.7 | -0.38816 | 2250 | 5736 |
| 27111 | 0.61 | -0.71 | -0.38816 | 2250 | 5736 |
| 27112 | 0.61 | -0.69 | -0.38816 | 2250 | 5736 |
| 27114 | 0.61 | 0 | 2250 | 5736 | |
| 27116 | -0.58 | 0.2 | -0.38816 | 2250 | 5736 |
| 27124 | -0.7 | 0.24 | -0.38816 | 2250 | 5736 |
| 27125 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27126 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27127 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27128 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27129 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27131 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27132 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27133 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27134 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27135 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27136 | -0.7 | 0.26 | -0.38816 | 2250 | 5736 |
| 27137 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27138 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27139 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27141 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27142 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27143 | -0.7 | 0.25 | -0.38816 | 2250 | 5736 |
| 27145 | -0.7 | 0.24 | -0.38816 | 2250 | 5736 |
| 27146 | -0.7 | 0.24 | -0.38816 | 2250 | 5736 |
| 27148 | -0.7 | 0.24 | -0.38816 | 2250 | 5736 |
| 27149 | -0.7 | -0.09 | 0.45743 | 2250 | 5736 |
| 27150 | -0.7 | -0.12 | 0.45743 | 2250 | 5736 |
| 27152 | -0.64 | -0.18 | 0.45743 | 2250 | 5736 |
| 27153 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27154 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27155 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27156 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27157 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27160 | -0.64 | -0.18 | 0.45743 | 2250 | 5736 |
| 27161 | -0.64 | -0.18 | 0.45743 | 2250 | 5736 |
| 27162 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27163 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27165 | -0.64 | -0.19 | 0.45743 | 2250 | 5736 |
| 27166 | -0.64 | 0.18 | -0.38816 | 2250 | 5736 |
| 27167 | -0.64 | 0.22 | -0.38816 | 2250 | 5736 |
| 27168 | -0.64 | 0.23 | -0.38816 | 2250 | 5736 |
| 27169 | -0.64 | 0.23 | -0.38816 | 2250 | 5736 |
| 27170 | -0.64 | 0.23 | -0.38816 | 2250 | 5736 |
| 27171 | -0.64 | 0.23 | -0.38816 | 2250 | 5736 |
| 27172 | -0.64 | -0.35 | 0.45743 | 2250 | 5736 |
| 27175 | -0.7 | -0.78 | 0.45743 | 2250 | 5736 |
| 27176 | -0.7 | -0.72 | 0.45743 | 2250 | 5736 |
| 27177 | -0.7 | -0.7 | 0.45743 | 2250 | 5736 |
| 27178 | -0.7 | -0.69 | 0.45743 | 2250 | 5736 |
| 27179 | -0.7 | -0.67 | 0.45743 | 2250 | 5736 |
| 27180 | -0.7 | -0.67 | 0.45743 | 2250 | 5736 |
| 27181 | -0.7 | -0.66 | 0.45743 | 2250 | 5736 |
| 27182 | -0.7 | -0.72 | 0.45743 | 2250 | 5736 |
| 27183 | -0.7 | -0.68 | 0.45743 | 2250 | 5736 |
| 27184 | -0.7 | -0.63 | 0.45743 | 2250 | 5736 |
| 27186 | -0.7 | 0.72 | -0.38816 | 2250 | 5736 |
| 27187 | -0.7 | 0.71 | -0.38816 | 2250 | 5736 |
| 27188 | -0.7 | 0.71 | -0.38816 | 2250 | 5736 |
| 27189 | -0.7 | 0.7 | -0.38816 | 2250 | 5736 |
| 27190 | -0.7 | 0.69 | -0.38816 | 2250 | 5736 |
| 27191 | -0.7 | 0.69 | -0.38816 | 2250 | 5736 |
| 27192 | -0.7 | 0.68 | -0.38816 | 2250 | 5736 |
| 27193 | -0.7 | 0.69 | -0.38816 | 2250 | 5736 |
| 27194 | -0.7 | 0.7 | -0.38816 | 2250 | 5736 |
| 27195 | -0.7 | 0.69 | -0.38816 | 2250 | 5736 |
| 27113 | 0.61 | 0.68 | 0.45743 | 2250 | 5736 |
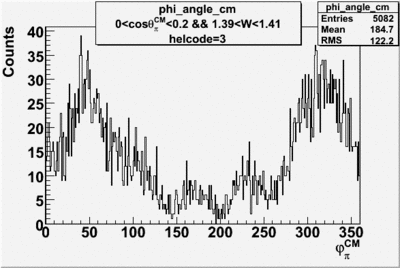
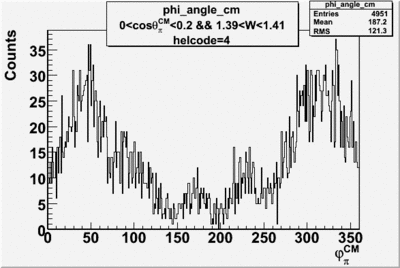
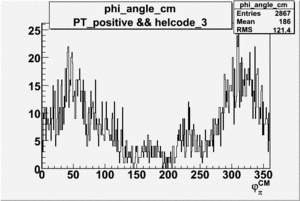
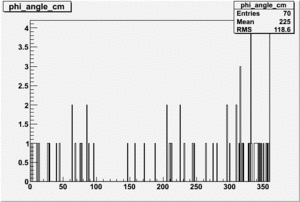
Phi_Angle_CM_Frame for Positive and Negative Target Polarization
| Helcode # | Negative Target Polarization | Positive Target Polarization |
| 1 h>0 | 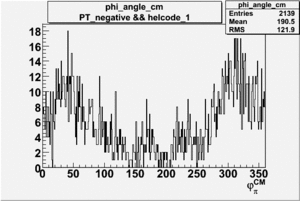 |
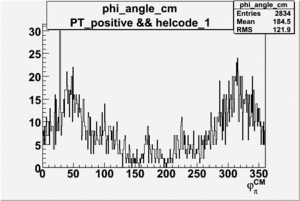
|
| 2 h<0 | 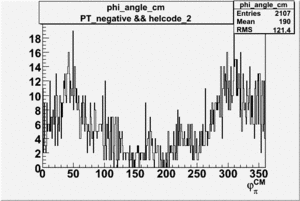 |
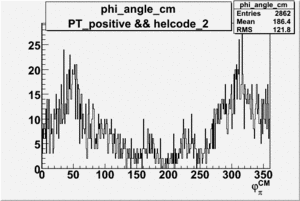
|
| 3 | 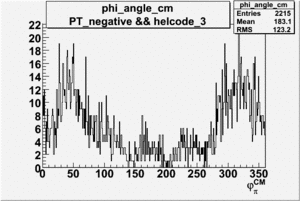 |
|
| 4 | 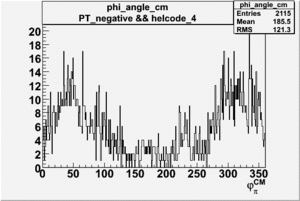 |
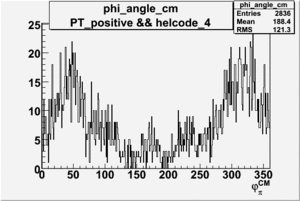
|
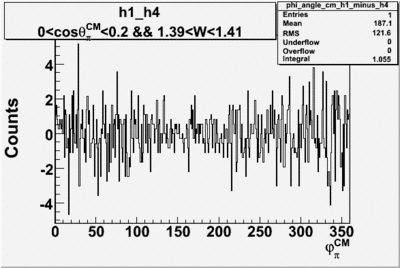
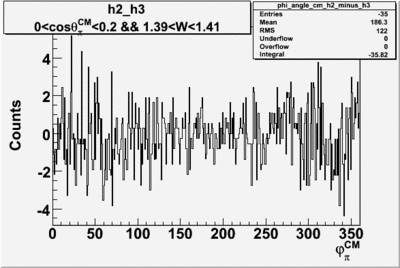
Helicity Difference Plots for Pt>0 and Pt<0
| Helicity Difference | Negative Target Polarization | Positive Target Polarization |
| h1-h4 | 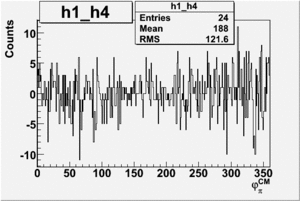 |
|
| h3-h2 |  |
|
5-01-2009
Target and Beam Polarization are positive
| Helcode # | Negative Beam Torus | Positive Beam Torus |
| 1 h>0 |  |
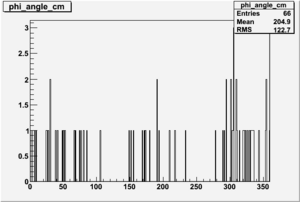
|
| 2 h<0 | 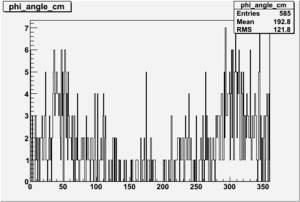 |
|
| 3 | 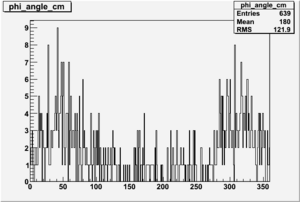 |
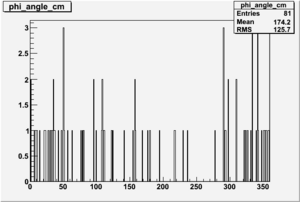
|
| 4 | 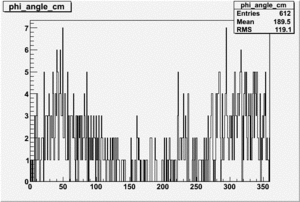 |
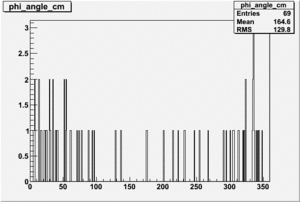
|
Target Polarization Positive and Beam Polarization Negative
| Helcode # | Negative Beam Torus | Positive Beam Torus |
| 1 h>0 | No Data | 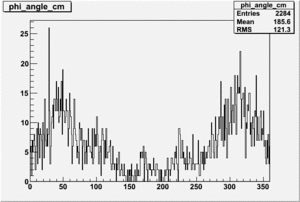
|
| 2 h<0 | No Data | 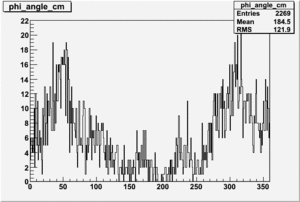
|
| 3 | No Data | 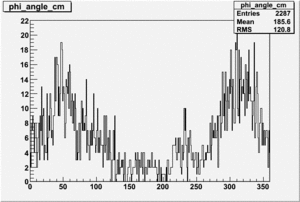
|
| 4 | No Data | 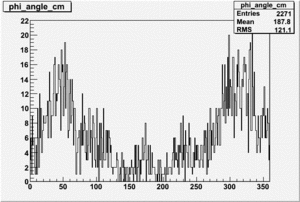
|
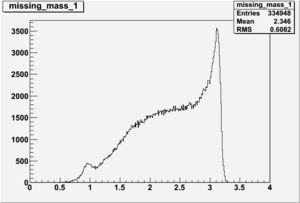
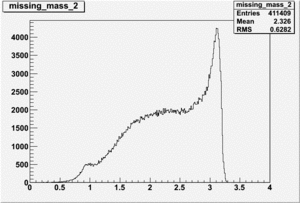
Inclusive Histograms For Invariant Mass
Invariant Mass Histograms for Each Sector
All helicities
Helcode=1
3/20/09
1.) plot Wdiff
2.) plot PE using Osipenko/Josh cuts
06/6/09
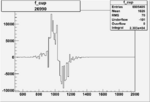
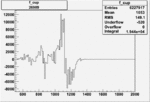
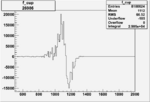
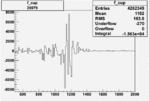
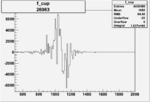
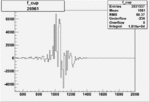
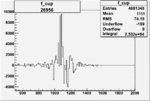
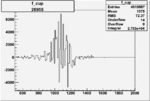
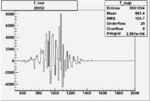
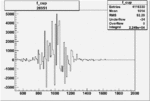
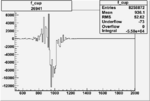
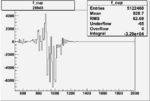
Invariant Mass
Difference of Invariant Mass for two differnet cuts:
| Run Number | EC Cuts+ requiring pion | OSI Cuts | EC Cuts | |
| 26991 | 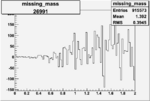 |
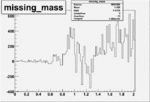 |
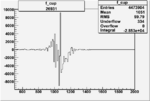
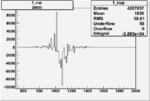
On x-axis of FC difference between the "+" heliciy FC and the "-" helicity FC.
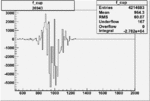
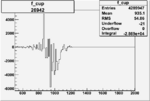
| Run Number | W difference | FC difference | End of Run sum |
| 26991 |  |
150px | 0.00354260538 0.00001050400 |
| 26994 | 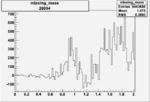 |
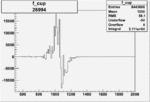 |
0.00367 0.00001088263 |
| 26993 | 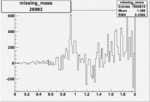 |
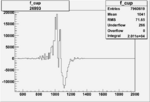 |
0.002559223 0.00001120796 |
| 26992 | 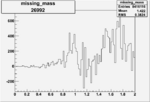 |
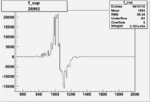 |
0.0037810.00001090044 |
| 26990 | 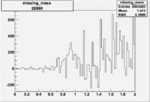 |
 |
0.003319863210.00001203387 |
| 26989 | 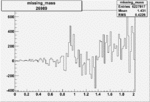 |
 |
0.003038415570.00001267152 |
| 26988 | 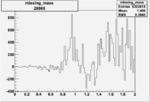 |
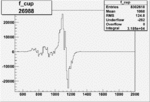 |
0.003801692430.00001097470 |
| 26987 | 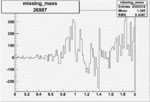 |
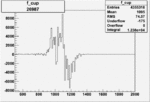 |
0.002801632400.00001515270 |
| 26986 | 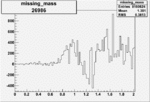 |
 |
0.003528826160.00000048897 |
| 26979 | 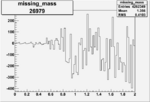 |
 |
-0.003731041260.00001531706 |
| 26965 | 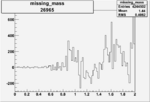 |
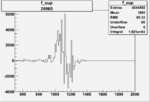 |
0.004541404390.00001534923 |
| 26964 | 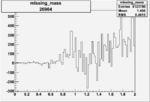 |
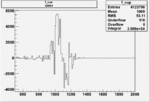 |
0.005094342830.00001557226 |
| 26963 | 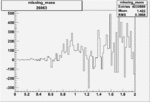 |
 |
0.004052851550.00001575049 |
| 26961 | 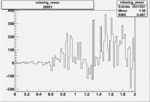 |
 |
0.00507456102 0.00001682744 |
| 26959 | 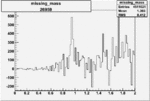 |
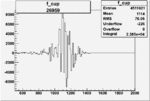 |
0.005237616940.00001488890 |
| 26958 | 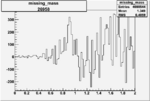 |
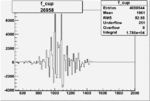 |
0.00444205478 0.00001577510 |
| 26956 | 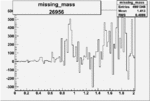 |
 |
0.005044016200.00001565456 |
| 26955 | 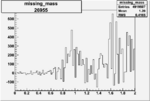 |
 |
0.005599138290.00001425736 |
| 26954 | 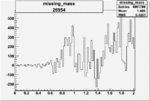 |
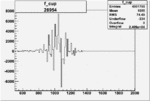 |
0.004946907280.00001443107 |
| 26953 | 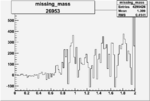 |
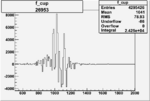 |
0.005625984480.00001525797 |
| 26952 | 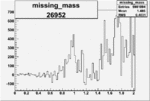 |
 |
0.004852958340.00001293033 |
| 26951 | 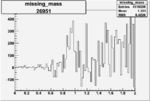 |
 |
0.005454859060.00001558637 |
| 26947 | 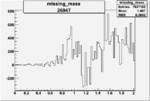 |
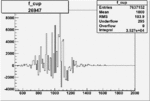 |
0.004656971600.00001144285 |
| 26945 | 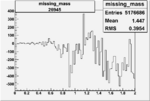 |
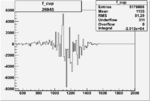 |
-0.00556456389 0.00001389870 |
| 26943 | 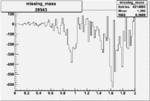 |
 |
-0.006561094720.00001540342 |
| 26942 | 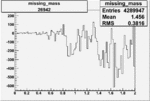 |
 |
-0.006691691060.00001526771 |
| 26941 | 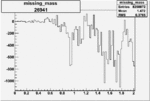 |
 |
-0.006764604170.00001100372 |
| 26940 | 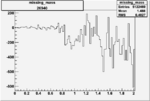 |
 |
-0.00643597022 0.00001397207 |
| 26939 | 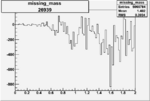 |
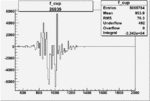 |
-0.006500474560.00001405001 |
| 26938 | 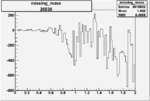 |
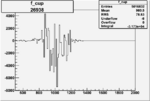 |
-0.006326303130.00001411839 |
| 26937 | 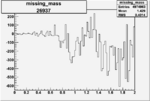 |
 |
-0.005331697940.00001417768 |
| 26934 |  |
 |
-0.006014971560.00001422560 |
| 26933 |  |
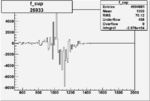 |
-0.005865405000.00001414938 |
| 26932 | 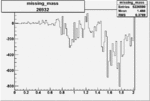 |
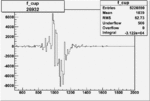 |
-0.005876871920.00001383218 |
| 26931 | 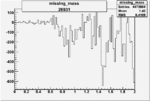 |
 |
-0.006302325660.00001495053 |
| 26930 | 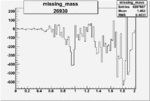 |
 |
-0.006842050100.00001541629 |
| 26929 | 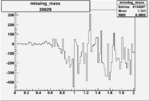 |
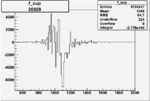 |
-0.005216669440.00001555256 |
| 26928 | 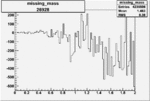 |
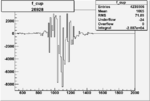 |
-0.006821853160.00001536552 |
| 26927 | 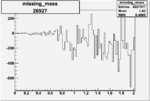 |
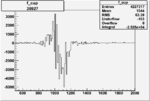 |
-0.005996474830.00001538040 |
| 26926 | 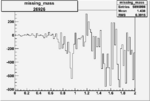 |
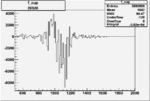 |
-0.006499323780.00001302331 |
| 26925 | 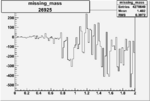 |
 |
-0.005754640990.00001530219 |
| 27079 |  |
 |
-0.010258694700.00002322298 |
| 27078 |  |
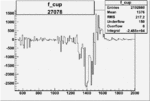 |
-0.011280362660.00002135573 |
| 27075 | 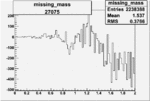 |
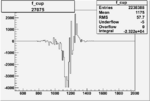 |
-0.010374430170.00002113646 |
| 27109 | 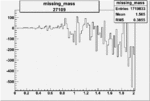 |
 |
-0.008768874650.00002411331 |
| 27107 |  |
 |
0.010445307520.00002188631 |
| 27116 |  |
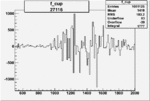 |
0.001732429540.00003084415 |
| 27112 | 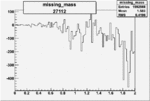 |
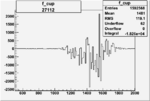 |
-0.011426827610.00002505827 |
| 27111 | 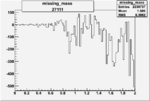 |
 |
-0.008727608860.00002117268 |
| 27128 |  |
 |
0.001934417560.00002177434 |
| 27127 |  |
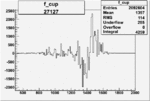 |
0.002157120980.00002186032 |
| 27124 | 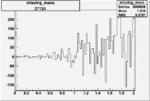 |
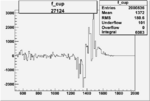 |
0.003090708080.00002235601 |
| 27139 | 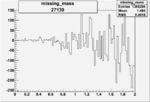 |
 |
-0.002888624440.00002683943 |
| 27138 |  |
 |
-0.000616086340.00002396188 |
| 27137 |  |
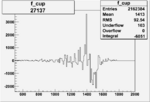 |
-0.002750667780.00002150471 |
| 27136 | 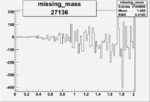 |
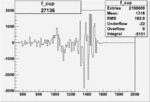 |
-0.002394232370.00002151354 |
| 27134 | 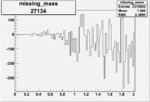 |
 |
-0.003555814340.00002051114 |
| 27133 |  |
 |
-0.002521937240.00002181781 |
| 27132 |  |
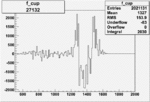 |
0.000963322020.00002224348 |
| 27143 | 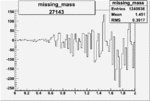 |
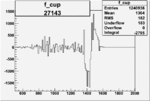 |
-0.002104862780.00002838738 |
| 27141 | 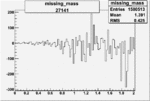 |
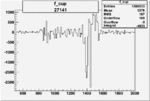 |
-0.003019905560.00002515365 |
| 27160 | 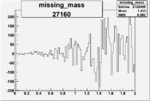 |
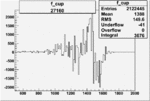 |
0.001712647440.00002170610 |
| 27161 | 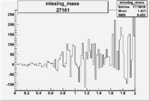 |
 |
0.002242873040.00002374488 |
| 27162 |  |
 |
0.001188444450.00002480837 |
| 27166 |  |
 |
0.000772370060.00002247548 |
| 27167 |  |
 |
0.002160412810.00002188669 |
| 27168 |  |
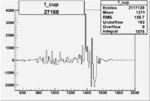 |
0.000836506250.00002173328 |
| 27170 | 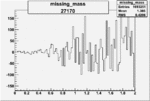 |
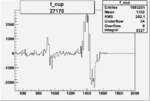 |
0.001512475110.00002430185 |
| 27175 | 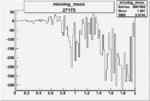 |
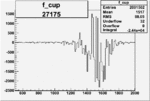 |
-0.012174856680.00002235229 |
| 27176 | 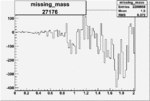 |
 |
-0.010496351670.00002127680 |
| 27177 | 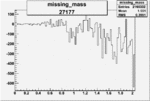 |
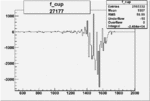 |
-0.011474452880.00002149007 |
| 27179 |  |
 |
-0.010253401530.00002206557 |
| 27180 |  |
 |
-0.008682724180.00002169057 |
| 27181 |  |
 |
-0.009733241880.00002374079 |
| 27182 |  |
 |
-0.009838624600.00002233926 |
| 27183 | 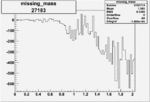 |
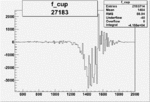 |
-0.019016152520.00002135060 |
| 27186 | 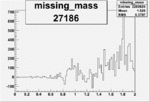 |
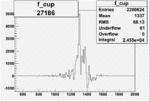 |
0.010790906350.00002093983 |
| 27187 |  |
 |
0.011555190760.00002157075 |
| 27188 |  |
 |
0.010868986050.00002089859 |
| 27190 | 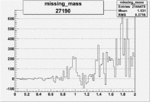 |
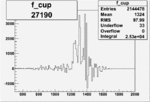 |
0.011812664900.00002159430 |
| 27192 | 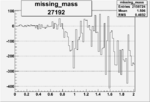 |
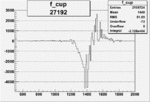 |
-0.009884596370.00002151795 |
| 27193 | 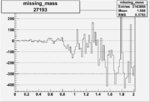 |
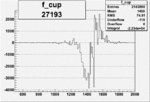 |
-0.010476430360.00002159742 |
| 27194 | 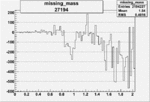 |
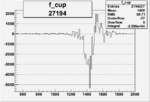 |
-0.011855200030.00002134810 |
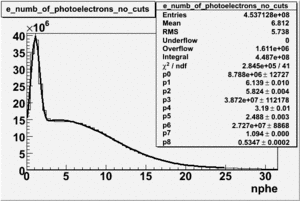
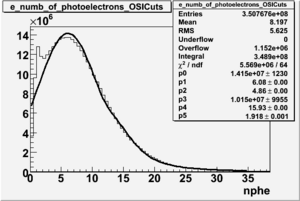
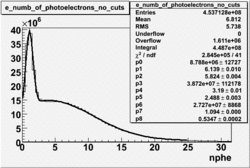
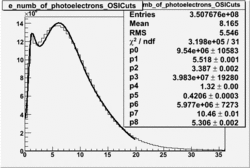
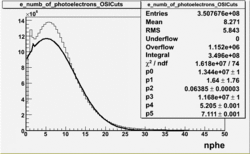
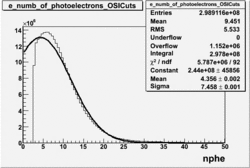
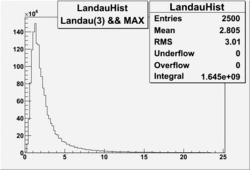
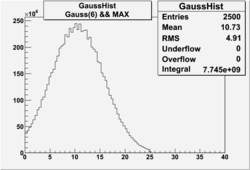
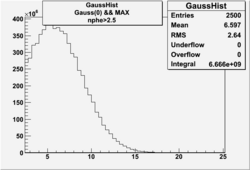
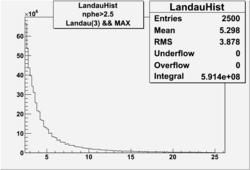
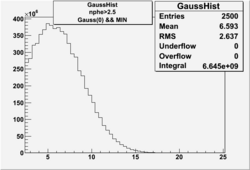
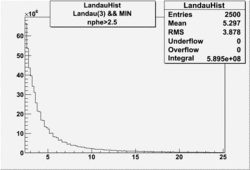
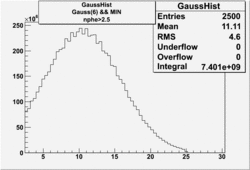
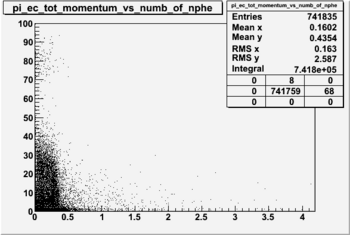
NPHE
To find out pion contamination in the electron sample i used Osipenko geometrical cuts. The number of photoelectrons before and after osipenko cuts are shown below:
| No cuts | OSI Cuts |
For different fits:
06/11/09
Pion Contamination
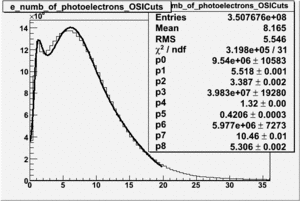
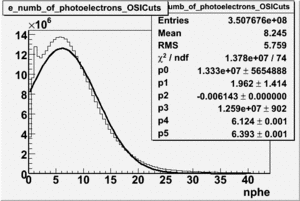
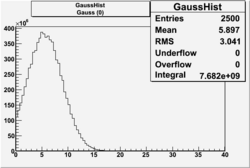
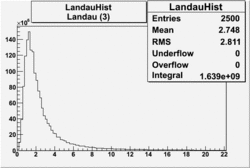
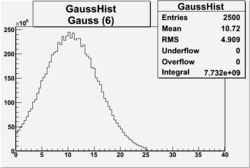
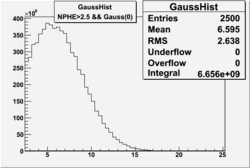
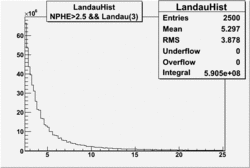
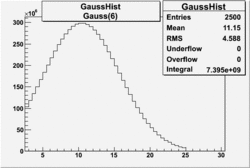
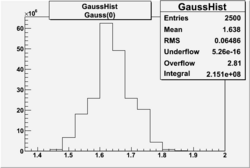
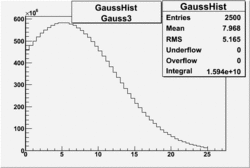
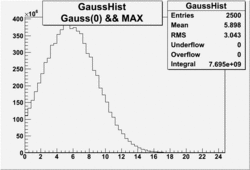
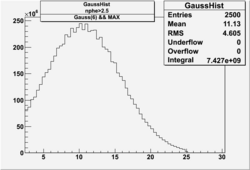
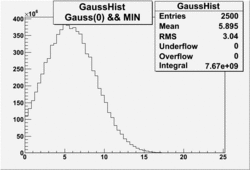
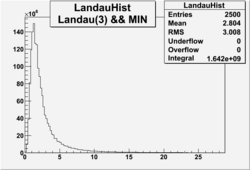
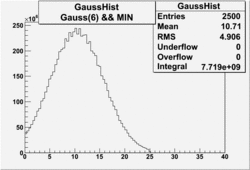
| No cuts | OSI Cuts (Gauss(0)+Landau(3)+Gauss(6)) | OSI Cuts (Gauss(0)+Gauss(3)) | OSICuts + NPHE>2.5 (Gauss) |
OSICuts (Gauss(0)+Landau(3)+Gauss(6))
Assuming that the two gaussians represent number of photoelectrons and landau number of photons produced by high energy pions, the ratio of number of pions over the sum of electrons and pion in the electron candidate sample can be calculated in the following way:
- OSIcut
- OSICut+NPHE>2.5
OSICuts (Gauss(0)+Gauss(3))
In case of only two gaussians, the number of photoelectrons produced by pions is described by Gauss(0) and the number of photoelectrons created by electrons is Gauss(3). Pion contamination is calculated below for both cases, without and with NPHE>2.5 cut.
- OSICut
- With NPHE>2.5 Cut
Number of Events after NPHE>2.5 Cut
=
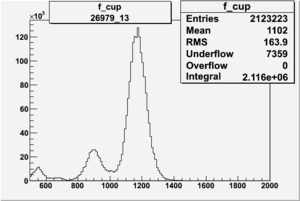
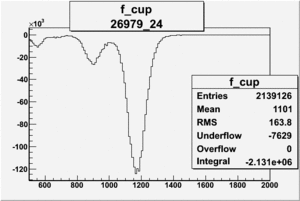
Counts in FCup
6/12/09
1.) Improve Chi^2 in NPe fits.
2.) Calculate uncertainty in pion contamination measurement by changing mean and widths according to fit error.
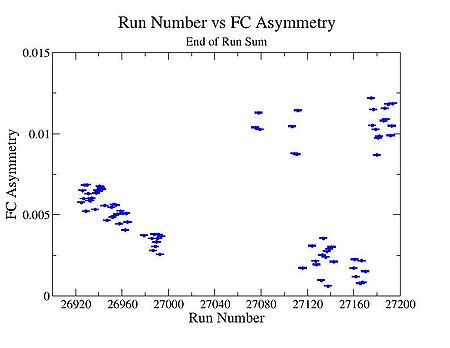
3.) Pulse pair FC asymmetry, and End of Run accumulated FC asym.
pulse pair
End of Run sum
4.) Determine semi-inclusive statistic as function of X
Uncertainty in Pion Contamination
Maximum
Minimum
Pion Contamination
It appears that pion contamination in electron sample is 9.63 % 0.01 % before nphe cut and after nphe>2.5 cut contamination is about 4.029% 0.003.
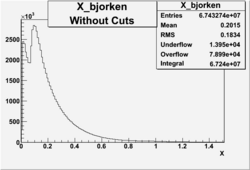
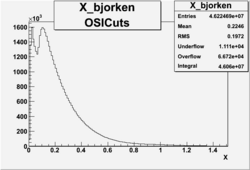
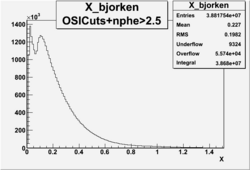
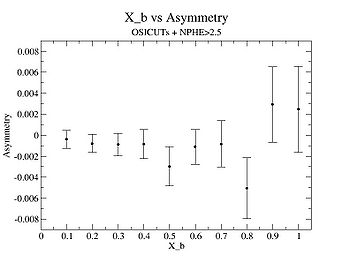
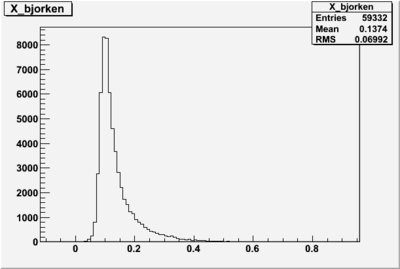
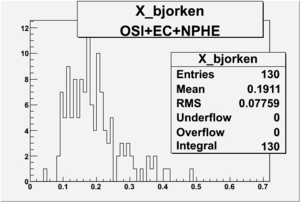
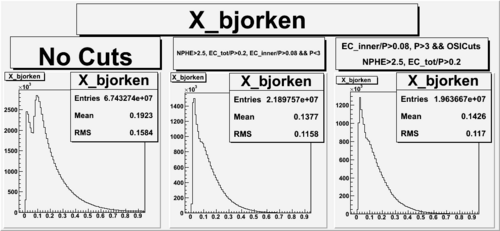
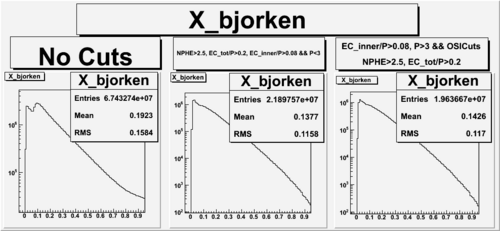
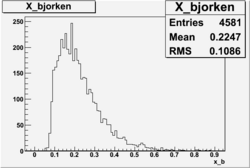
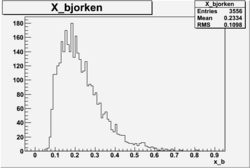
X_bjorken
- 1). alldataOSICuts_X.root - OSICuts applied.
- 2). alldataOSICuts_X_epx.root - OSICuts applied and electron and pion are required.
- 3). alldataOSICuts_X_epxnphe.root - OSICut and nphe>2.5 cuts applied and electron and pion are required.
- 4). alldataX_epxwithoutcuts - No cuts, electron and pion required.
- Number of Events after cuts
| No Cuts | OSI Cuts | OSI+NPHE>2.5 Cuts |
| 68.5 % | 57.5 % |
- Error Calculation
Tthe error in the asymmetry measurement would be
| X_b | X_b Asymmetry | Error | ||
| 0.1 | 0.00087251693 | |||
| 0.2 | 0.0008507530911145 | |||
| 0.3 | 1.0691459e-03 | |||
| 0.4 | 0.0014004231 | |||
| 0.5 | 0.0018665742 | |||
| 0.6 | 0.0016477095 | |||
| 0.7 | 0.0022190018 | |||
| 0.8 | 0.00291609 | |||
| 0.9 | 0.003592967 | |||
| 1 | 0.0040928449 |
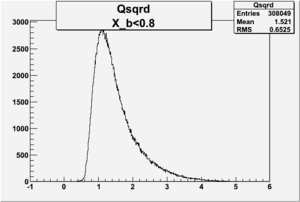
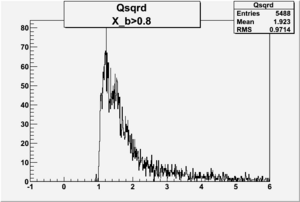
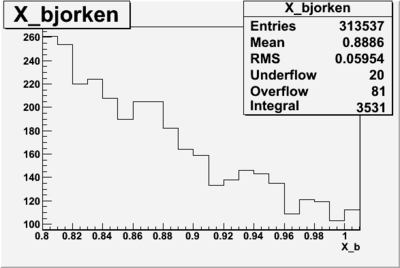
I am pretty sure X_{BJ} > 0.8 is not possible with our data set
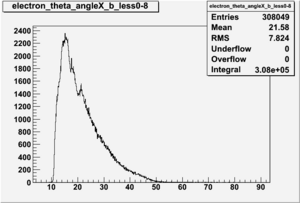
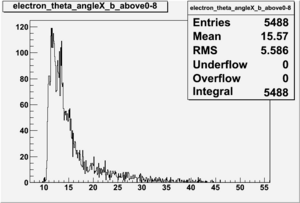
Electron theta angle and cuts
- electron theta angle for different X_b
X_b when
Number of Events for X_b>0.8
plot the vertex of the above hits with X>0.8
change below to log plots so we can see where XBj stops
10/23/09
After Months of working on detectors and writing thesis proposal it is now time to start doing some physics.
1.) Determine how pion contamination uncertainty changes when you change fit parameters by 1 S.D., 2 S.D., and 3 S.D.
2.) FC asymm plots
3.) Vertex plot for X > 0.8 events.
4.) Now that we have good electron cuts. Plot statistics for Pion cuts.
5.) After pion cuts we start looking add paddle efficiencies so we can subtract sem-inclusive rates using individual paddles but opposite magnetic fields.
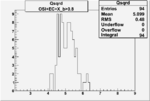
Xbjorken
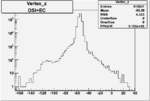
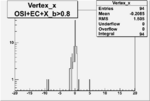
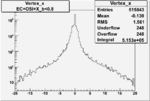
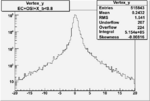
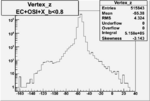
Vertex plot for X > 0.8 events and others
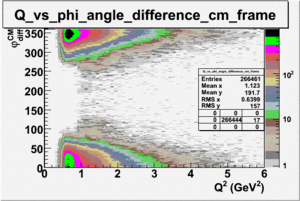
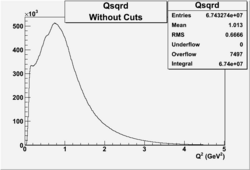
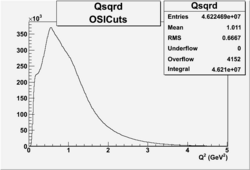
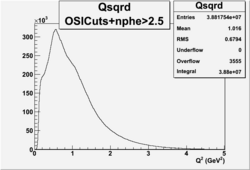
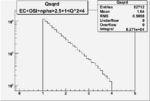
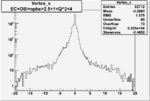
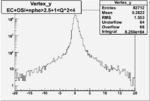
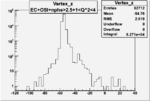
I chose 1<Q^2<4 cut because we used it to plot phi angle in cm frame vs relative rate to compare with the results in paper.
| Cuts | X_bjorken | Vertex X | Vertex Y | Vertex Z | |
| OSI Cuts + EC Cuts | 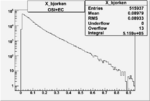 |
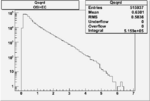 |
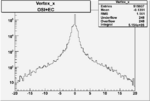 |
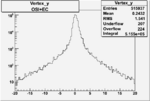 |
|
| OSI Cuts + EC Cuts + X_b>0.8 | 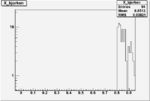 |
 |
 |
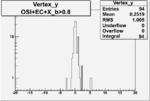 |
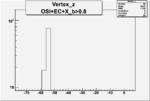
|
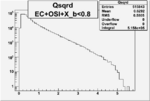
| OSI Cuts + EC Cuts + X_b<0.8 | 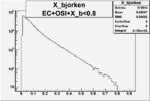 |
 |
 |
 |
|
| OSI Cuts + EC Cuts + 1<Q^2<4 | 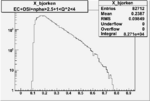 |
 |
 |
 |
|
| Cuts | The scattered electron energy | electron scattering angle |
 |
| |
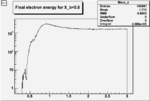 |
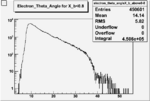
|
FC Asymmetry
FC Asymmetry plot using the following method : End of Run sum
Pion contamination
Determine how pion contamination uncertainty changes when you change fit parameters by 1 S.D., 2 S.D., and 3 S.D.
Pion contamination(3 S.D.) in electron sample is 9.645 % 0.025%.
It doesnt really change from using 1 S.D.
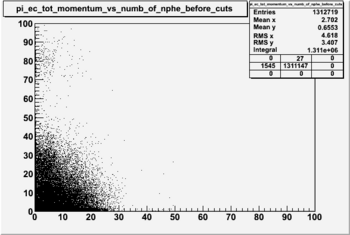
Pion Statistics
Before and after cuts the plot of EC_tot/P vs nphe(for pions)
10/30/09
After Months of working on detectors and writing thesis proposal it is now time to start doing some physics.
1.) Determine how pion contamination uncertainty changes when you change fit parameters by 1 S.D., 2 S.D., and 3 S.D.
2.) Do pulse pair FC asymm plot
3.) Check program's calculation of event with X > 0.8 events. and compare to similar event with X < 0.8
4.) Use statistics for Pion cuts to estimate SIDIS statistical error -vs- Xbj
5.) After pion cuts we start looking add paddle efficiencies so we can subtract sem-inclusive rates using individual paddles but opposite magnetic fields.
1.)
1.) Determine how pion contamination uncertainty changes when you change fit parameters by 1 S.D., 2 S.D., and 3 S.D.
In case of 10 S.D. : &&
3.)
I suspect the X_b >0.8 event below are pions mis-identified as electrons To figure out. Write down event number for events below as well as run number and file name. The use path length and Scintillator TDC time to determine beta under assumption that particle is a pion. Does the momentum and energy make sense? Download the cooked data file from JLab for these events so we can use CED to look at them and bosdump to look at the reconstruction.
- 1
Ebeam=5736
IBeam=4.2
ITorus=2248
ITarg=122
BeamPol=0.71
TargetPol=-0.67
BadRun=0
Target=18
PolPlate=0
Version=2
Prescalers:0:0:0:0:0:0:0
dump=13
W= 1.3504
Q= 4.78723
final electron energy= 2.6823
initial electron energy= 5.736
electron theta angle= 32.3896
- 2
Ebeam=5736
IBeam=4.2
ITorus=2248
ITarg=122
BeamPol=0.71
TargetPol=-0.67
BadRun=0
Target=18
PolPlate=0
Version=2
Prescalers:0:0:0:0:0:0:0
dump=13
W= 1.41085
Q= 5.1186
final electron energy= 2.41678
initial electron energy= 5.736
electron theta angle= 35.3749
- 3
Ebeam=5736
IBeam=4.2
ITorus=2248
ITarg=122
BeamPol=0.71
TargetPol=-0.67
BadRun=0
Target=18
PolPlate=0
Version=2
Prescalers:0:0:0:0:0:0:0
dump=13
W= 1.2575
Q= 4.83661
final electron energy= 2.7851
initial electron energy= 5.736
electron theta angle= 31.9378
- 1
W= 3.07188 Q= 0.229403 final electron energy= 1.0543 initial electron energy= 5.736 electron theta angle= 11.177
- 2
hit return for next event, q to quit: W= 2.48202 Q= 1.382 final electron energy= 2.18587 initial electron energy= 5.736 electron theta angle= 19.1106
- 3
hit return for next event, q to quit: W= 2.92788 Q= 0.274895 final electron energy= 1.49046 initial electron energy= 5.736 electron theta angle= 10.2878
4.)
| && | && | && | && |
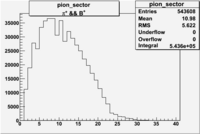 |
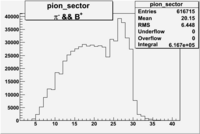 |
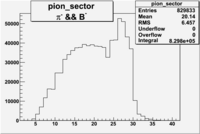 |
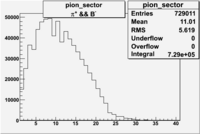
|
1-12-09
Root files: alldatasector26990_4.root, alldatasector26990_5.root, alldatasector27113_4.root, alldatasector27113_5.root.
9/13/09
1.) Determine how pion contamination uncertainty changes when you change fit parameters.
| Fit parameters | Pion Contamination |
| 1 S. D. | 9.63 % 0.01 % |
| 3 S. D. | 9.645 % 0.025 % |
| 10 S. D. | 9.446 % 0.0233 % |
2.) Do pulse pair FC asymm plot
I did it for one file(dst27113_00.B00) and it was zero.
3.) Check program's calculation of event with X > 0.8 events. and compare to similar event with X < 0.8
I suspect the X_b >0.8 event below are pions mis-identified as electrons To figure out. Write down event number for events below as well as run number and file name. The use path length and Scintillator TDC time to determine beta under assumption that particle is a pion. Does the momentum and energy make sense? Download the cooked data file from JLab for these events so we can use CED to look at them and bosdump to look at the reconstruction.
Calculation is right, need to check CED, but dont have it on daq.
4.) Use statistics for Pion cuts to estimate SIDIS statistical error -vs- Xbj
Insert table with X bj, number of reconstructed pions, statistical error.
no cut
root alldatasector27113_5_1.root
root [9] .p X_bjorken->GetBinError(2); (const Double_t)3.50713558335003626e+01 root [10] .p X_bjorken->GetBinContent(2); (const Double_t)1.23000000000000000e+03
root [11] .p X_bjorken->GetBinContent(3); (const Double_t)1.85200000000000000e+03 root [12] .p X_bjorken->GetBinError(3); (const Double_t)4.30348695827000256e+01
root [13] .p X_bjorken->GetBinError(4); (const Double_t)3.06431068920891256e+01 root [14] .p X_bjorken->GetBinContent(4); (const Double_t)9.39000000000000000e+02
root [15] .p X_bjorken->GetBinContent(5); (const Double_t)3.68000000000000000e+02 root [16] .p X_bjorken->GetBinError(5); (const Double_t)1.91833260932508765e+01
(const Double_t)1.91833260932508765e+01 root [17] .p X_bjorken->GetBinError(6); (const Double_t)1.14017542509913792e+01 root [18] .p X_bjorken->GetBinContent(6); (const Double_t)1.30000000000000000e+02
root [19] .p X_bjorken->GetBinContent(7); (const Double_t)4.10000000000000000e+01 root [20] .p X_bjorken->GetBinError(7); (const Double_t)6.40312423743284853e+00
root [21] .p X_bjorken->GetBinError(8); (const Double_t)3.74165738677394133e+00 root [22] .p X_bjorken->GetBinContent(8); (const Double_t)1.40000000000000000e+01
root [23] .p X_bjorken->GetBinContent(9); (const Double_t)5.00000000000000000e+00 root [24] .p X_bjorken->GetBinError(9); (const Double_t)2.23606797749978981e+00
root [25] .p X_bjorken->GetBinError(10); (const Double_t)1.41421356237309515e+00 root [26] .p X_bjorken->GetBinContent(10); (const Double_t)2.00000000000000000e+00
with cut
root alldatasector27113_5.root
root [3] .p X_bjorken->GetBinError(2); (const Double_t)2.89827534923788761e+01 root [4] .p X_bjorken->GetBinContent(2); (const Double_t)8.40000000000000000e+02
root [5] .p X_bjorken->GetBinError(3); (const Double_t)3.77491721763537456e+01 root [6] .p X_bjorken->GetBinContent(3); (const Double_t)1.42500000000000000e+03
root [7] .p X_bjorken->GetBinError(4); (const Double_t)2.82488937836510701e+01 root [8] .p X_bjorken->GetBinContent(4); (const Double_t)7.98000000000000000e+02 root [9] .p X_bjorken->GetBinError(5); (const Double_t)1.81107702762748346e+01 root [10] .p X_bjorken->GetBinContent(5); (const Double_t)3.28000000000000000e+02
root [11] .p X_bjorken->GetBinError(6); (const Double_t)1.04880884817015154e+01 root [12] .p X_bjorken->GetBinContent(6); (const Double_t)1.10000000000000000e+02
root [13] .p X_bjorken->GetBinError(7); (const Double_t)6.24499799839839831e+00 root [14] .p X_bjorken->GetBinContent(7); (const Double_t)3.90000000000000000e+01
root [15] .p X_bjorken->GetBinError(8); (const Double_t)3.60555127546398912e+00 root [16] .p X_bjorken->GetBinContent(8); (const Double_t)1.30000000000000000e+01
root [17] .p X_bjorken->GetBinError(9); (const Double_t)1.73205080756887719e+00 root [18] .p X_bjorken->GetBinContent(9); (const Double_t)3.00000000000000000e+00
root [19] .p X_bjorken->GetBinError(10); (const Double_t)0.00000000000000000e+00 root [20] .p X_bjorken->GetBinContent(10); (const Double_t)0.00000000000000000e+00
root [21] .p X_bjorken->GetBinError(11); (const Double_t)0.00000000000000000e+00 root [22] .p X_bjorken->GetBinContent(11); (const Double_t)0.00000000000000000e+00
root [23] .p X_bjorken->GetBinError(12); (const Double_t)0.00000000000000000e+00 root [24] .p X_bjorken->GetBinContent(12); (const Double_t)0.00000000000000000e+00
| Error | |
| 0.1 | 0.034503278 |
| 0.2 | 0.026490647 |
| 0.3 | 0.035399616 |
| 0.4 | 4.255675067 |
| 0.5 | 0.095346259 |
| 0.6 | 0.160128154 |
| 0.7 | 0.277350098 |
| 0.8 | 0.577350269 |
5.) After pion cuts we start looking add paddle efficiencies so we can subtract sem-inclusive rates using individual paddles but opposite magnetic fields.
Your B-field sign change does effect paddle distribution?
The table below represents the distribution of electrons and pions on the scintillator paddles using the reaction
e(p/d,e')\pi X
| File number | electron | electron | ||
| 26990, B<0 | 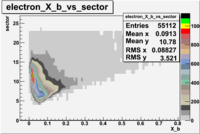 |
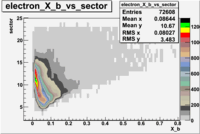 |
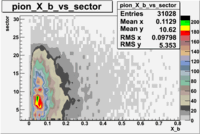 |
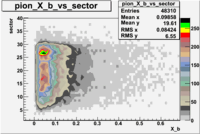
|
| 27113, B>0 | 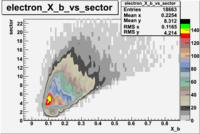 |
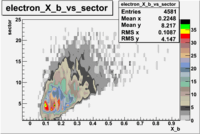 |
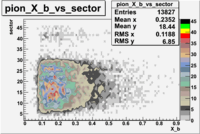 |
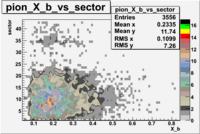
|
- 1.) 7< Sector_paddle <11 - (B>0, ) - 15.3% && (B<0, ) - 10.9%
- 2.) 25< Sector_paddle <29 - (B>0, ) - 7.775% && (B<0, ) - 10.97%
&&
Using Two runs: 26990(NH3, -2250) and 27124(ND3, +2250)
root files: alldatasector27124_4.root && alldatasector27124_5.root
root files: alldatasector26990_4.root && alldatasector26990_5.root
alldatasector26990_4.root
Pion Paddle Number
Negative Torus
Using runs with NH3 target
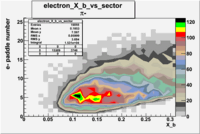
| Detected particles in the final state | Pion Paddle Number | X_b vs pion paddle number | Chosen pion paddle number |
| && | 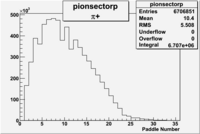 |
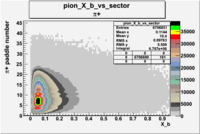 |
7 |
| && | 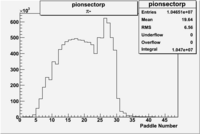 |
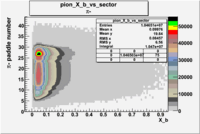 |
27 |
| Detected particles in the final state and chosen pion paddle number | paddle number | vs paddle number | vs paddle number when | vs paddle number when |
| && , | 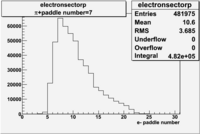 |
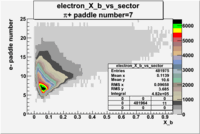 |
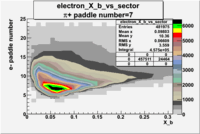 |
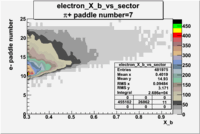
|
| && , | 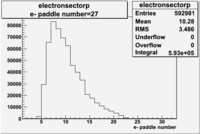 |
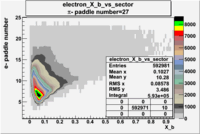 |
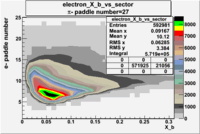 |
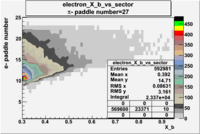
|
Positive Torus
Using runs with NH3 target
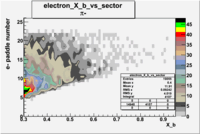
| Detected particles in the final state | X_b vs pion paddle number | Chosen pion paddle number |
| && | 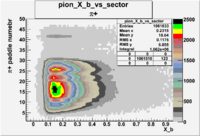 |
27 |
| && | 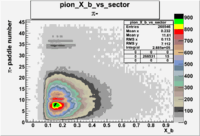 |
7 |
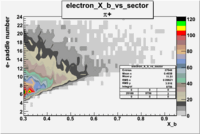
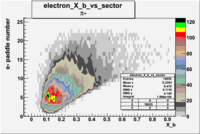
| Detected particles in the final state and chosen pion paddle number | paddle number | vs paddle number | vs paddle number when | vs paddle number when |
| && , | 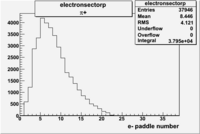 |
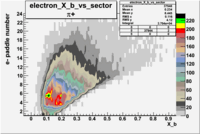 |
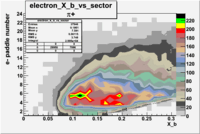 |
|
| && , | 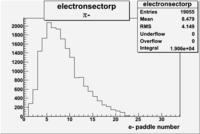 |
 |
 |
|
1/15/2010
1.) How different are the kinematics when Pion in paddle 27 and electron in paddle 11 for the two different B-field
2.) Rate differences when B>0 && \pi+ in paddle 27 && e- in paddle 8 to B<0 && \pi- in paddle 27 && e- in paddle 11
3.) Do similar analysis for pi+ && B< 0
4.) Now we need to look at W. Selected SIDIS event for Xb > 0.3. plot W -vs- electron paddle number when pions are in paddle 27 and 7.
| root file | reaction |
| 1 | B<0, pi^-=27 && e^-=11 |
| 2 | B>0, pi^+=27 && e^-=11 |
| 3 | B>0, pi^+=27 && e^-=7 |
| 4 | B>0, pi^-=7 && e^-=11 |
| 5 | B<0, pi^+=7 && e-=11 |
1 Kinematics comparison
| Selection of paddle number and etc | W vs | |||