SPIM ComptonScattering Lab
The goal of this assignment is to compare the Compton Scattering Energy Differential cross-section () simulated in GEANT4 to the Klein-Nishina Formula using the material selected for the evaluation of GEANT4's photoelectric effect comparison.
Create histogram of dSigma/dE using Klein-Nishina formula.
A C-program is given below which will output the Electron Kinetic energy and the differential X-sect
/*
A program to calculate the Energy Differential Compton Scattering
Crosssection (d Sigma/ dE)
in units of barns using the Klein-Nachina formula.
pass electron min kinetic energy, Photon Energy and step size (Emin,Egamma,Estep)
*/
/* system include files for input and output*/
#include <stdio.h>
#include <stdlib.h>
#include <math.h>
main(int argc, char *argv[])
{
int i,loop;
int Emin,Estep,Egamma;
double Emax,Eelectron,PI,DSigma_DE,R,Bohr_Radius_sqrd,Eo_electron,s,psi;
PI=3.1415927;
Bohr_Radius_sqrd=2.817e-13*2.817e-13/1e-24;/*barns*/
if(argc==4)
{
/* convert the comand line string to an integer*/
Emin=atoi(argv[1]);
Egamma=atoi(argv[2]);
Estep=atoi(argv[3]);
}
else
{
fprintf(stderr,"Error you must enter 3 energies in eV on the comand line\n");
fprintf(stderr,"usage: DiffEXsect [electron min KE] [Incident photon E] [elecron KE step size]\n");
exit(1);
}
Eo_electron=511000; /* rest mass of electronin eV */
psi=Egamma/Eo_electron;
Emax=Egamma*2*psi/(1+2*psi); /* compton edge */
fprintf(stderr,"Calculating Differential Compton cross section (in barns/eV) for \n\t %d < E < %g in steps of %d and %d eV incident Photons\n",Emin,Emax,Estep,Egamma);
Eelectron=Emin-Estep;
for(i=Emin;i<=Emax; i+=Estep)
{
Eelectron+=Estep;
/* fprintf(stderr,"Eelectron=%g\n",Eelectron);*/
s=Eelectron/Egamma;
DSigma_DE=s*(s-2/psi)/(1-s);
DSigma_DE=DSigma_DE+s*s/(1-s)/(1-s)/psi/psi;
DSigma_DE=2.0+DSigma_DE;
DSigma_DE=PI*Bohr_Radius_sqrd*DSigma_DE/Eo_electron/psi/psi;
fprintf(stdout,"%g\t%g\n",Eelectron,DSigma_DE);
}
}
save the above file under the name "DiffEKleinNishina.c "
The makefile below can be used to compile the above program. Just copy the lines below into a file and save the file under the filename "Makefile". If your program and the Makefile and in the same subdirectory, then your program should compile by typing "make".
CC = gcc
DiffEXsect: DiffEKleinNishina.o
$(CC) $(CFLAGS) DiffEKleinNishina.o -lm -o DiffEXsect
DiffEKleinNishina.o: DiffEKleinNishina.c
$(CC) $(CFLAGS) -c DiffEKleinNishina.c
Sometimes, if you copy and paste from the browser you get control characters which cause the error message " *** missing separator.". Just delete everything in front of the $(CC) line and insert our own tab character.
Compile and run the program with the commands
make DiffEXsect
./DiffEXsect 100 8000 10 > DiffEXsect.dat
this will output the ejected electron kinetic energy (eV) and differential energy Xsection (barns/eV) to the file "DiffEXsect.dat" for an incident photon energy of 8 keV starting at the ejected electron energy of 100 eV in steps of 10 eV.
Create ROOT histogram
Now that you have a data file you will need to create a ROOT histogram so you can overlay the theory with the GEANT4 simulation.
This macro will convert the data file into a root file
copy the macro into a file called "DiffEXsect2root.C"
void DiffEXsect2root() {
struct evt_t {
Int_t Egamma;
Float_t Eelectron, dSigma_dE;
};
ifstream in;
in.open("DiffEXsect.dat");
evt_t evt;
Int_t nlines=0;
TFile *f = new TFile("DiffEXsect.root","RECREATE");
TTree *tree = new TTree("DiffEXsect","Compton Diff Xsect dSigma/dE");
tree->Branch("evt",&evt.Egamma,"event/I:Eelectron/F:dSigma_dE");
while(in.good()){
evt.Egamma=80000;
in >> evt.Eelectron >> evt.dSigma_dE;
/*
printf( " %d %f %f %f %f %f %f %f\n",evt.event, evt.KE, evt.pos[0], evt.pos[1], evt.pos[2], evt.mom[0], evt.mom[1], evt.mom[2] );
*/
nlines++;
tree->Fill();
}
tree->Print();
tree->Write();
in.close();
delete tree;
delete f;
}
you then execute the above function within root via:
.X DiffEXsect2root.C
assuming you called the date file DiffEXsect.dat and the above script filename "DiffEXsect2root.C"
Go ahead and quit root so you can restart fresh.
Now that you have the root tree in a file, load it into root
root DiffEXsect.root
the commands below will fill a histogram called "h222" with the data from the file
TTree *tree=DiffEXsect;
DiffEXsect->Draw("Eelectron >> h222(25,0,250)","dSigma_dE")
Note that if you change the incident photon energy you need to change the bins in "h222" above.
Also note: The above cross-section is for a single free electron. You need to multiply by the number of atomic electrons (Z) to get the Atomic compton cross-section.
GEANT program
Warning geant4.9.4 and more recent may not have LowEnergy model
Generate GEANT4 file using two different Compton Scattering Models (regular and LowEnergy).
Add G4EnergyCompton to your physics list
edit the physics list file and be sure the header file
#include "G4ComptonScattering.hh"
is included then go down to the gamma physics list
if (particleName == "gamma") {
// gamma
// ph->RegisterProcess(new G4PhotoElectricEffect, particle);
ph->RegisterProcess(new G4ComptonScattering, particle);
//ph->RegisterProcess(new G4GammaConversion, particle);
}
now change the stepping verbose so it outputs compton data
In the usual spot where values are written to an output file your if statement could read
if( fTrack->GetDefinition()->GetPDGEncoding()==22 && fStep->GetPostStepPoint()->GetProcessDefinedStep()->GetProcessName() == "compt" && fTrack->GetVolume()->GetName() =="Target")
{
PhotonEloss=InitialPhotonEnergy-fTrack->GetKineticEnergy();
//G4cout << " Photon " ;
//G4cout
outfile
<< PhotonEloss << " "
<< InitialPhotonEnergy << " "
<< fTrack->GetPosition().x()<< " "
<< fTrack->GetPosition().y()<< " "
<< fTrack->GetPosition().z()<< " "
<< fTrack->GetMomentum().x() << " "
<< fTrack->GetMomentum().y() << " "
<< fTrack->GetMomentum().z() << " "
<< G4endl;
}
Notice I have two new variables that are not in the photoelectric effect version from last time. YOU MAY NOTICE THAT GEANT4 DOESN'T CALCULATE THE PHOTON ENERGY LOSS (a.ka. ELECTRON KINETIC ENERGY) FOR THE ROUTINE "compt" SO YOU WILL NEED TO DO IT BY HAND
<< PhotonEloss << " " << InitialPhotonEnergy << " "
I set the InitialPhoton Energy when the tracking enters the world volume using the if statement
if( fTrack->GetVolume()->GetName() =="World")
{
if( fTrack->GetDefinition()->GetPDGEncoding()==22 && fTrack->GetTrackID()==1)
InitialPhotonEnergy=fTrack->GetKineticEnergy();
}
The above if statement happens before the if statment to see if stepping is in the Target volume!
Be sure to add the variables PhotonEloss and InitialPhotonEnergy to the include file include/ExN02SteppingVerbose.hh
Float InitialPhotonEnergy; Float PhotonEloss;
As always check the output by looking at events interactively on the screen. (Un-comment "G4cout" and comment "outfile" above)
If Using G4 version numerically smaller than 4.9.5 Add G4LowEnergyCompton to your physicslist
include the header file below if LowEnergy isn't defined
#include "G4LowEnergyCompton.hh"
Notice the tracking calls this process "LowEnCompton"
Add this process under the gamma particle (or just comment out the other ones):
if (particleName == "gamma") {
// gamma
pmanager->AddDiscreteProcess(new G4ComptonScattering);
}
Write up results
Write up the results in report similar to the photoelectric effect homework. The grading scheme will be
0 points /10 if you use a target atom with less than 3 energy levels (ie H, He, Li ...). This means all atoms should have "A" of Sodium(Na) or above.
8 points/10 if you supply a pdf file containing a graph with the results from theory and GEANT4.
9 points /10 if the above graph has the units of cross section.
10 points /10 if you provide a graph of the percent difference ((theory-GEANT4)/Theory).
NOTE: The Kinetic Energy histogram from GEANT is in units of counts and not barns. The cross-section is given by a ratio of (the scattered particle/solid angle) to (the incident particles/area). You are detecting particles in a solid angle of 4pi sterradians. To determine the number of incident particles/area think of the target as the beam of incident particles. Then you just need to know the target length (cm) and density(Atoms/cm^3) to determine how many particles you threw at each photon from the gun. You need to multiply that number by the number of times the gun fired to get the total number of incident particles per area. Then note that a barn is and you can convert the counts in the GEANT energy distribution to units of barns.
HINT: How to multiply or divide histograms in ROOT
Lets assume I have a root file called "Compton.root" with a Tree called "Compton"
Here are the root commands I used to multiply the counts in a KE distribution stored in a histogram called "C1" by 10 and store the result in a histogram called "C2"
root Compton.root
root [1] TTree *tree=Compton;
root [2] TH1F *C1=new TH1F("C1","C1",40,0,0.4);
root [3] TH1F *C2=new TH1F("C2","C2",40,0,0.4);
root [4] Compton->Draw("ke*1000 >> C1");
<TCanvas::MakeDefCanvas>: created default TCanvas with name c1
root [5] C2->Add(C1,10);
root [6] C2->Draw();;
My root session looked like this:
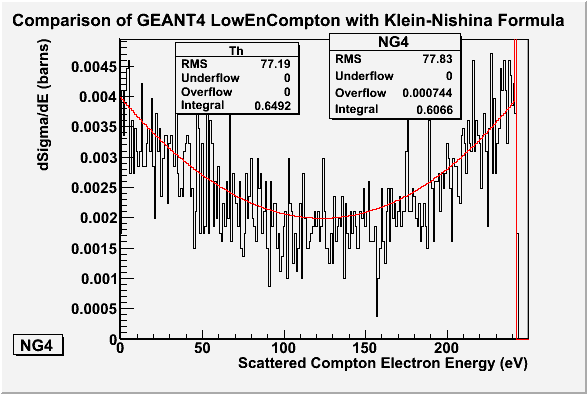
I used a 30cm long Argon gas and 10^7 incident photons.
root [1] TTree *Sim=Photo;
root [2] TH1F *G4=new TH1F("G4","G4",250,-0.5,249.5);
root [3] Sim->Draw("ke*1000000 >> G4");
root [4] TTree *Theory=DiffEXsect;
root [5] TH1F *Th=new TH1F("Th","Th",250,-0.5,249.5);
root [6] Theory->Draw("Eelectron >> Th","dSigma_dE");
root [7] TH1F *NG4=new TH1F("NG4","NG4",250,-0.5,249.5);
root [8] NG4->Add(G4,1.24e-4);
root [9] NG4->Draw();
root [10] Th->Draw("same");
My plot using 10 million 8 keV photons on Argon is shown below
HomeWork_Simulations_of_Particle_Interactions_with_Matter#Homework_8