Difference between revisions of "Run in GEMC"
| Line 111: | Line 111: | ||
<pre> | <pre> | ||
| − | split -d -l 3000 -a | + | split -d -l 3000 -a 3 LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut.LUND LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_ |
</pre> | </pre> | ||
| Line 117: | Line 117: | ||
<pre> | <pre> | ||
| − | prename 's/(LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_\d{ | + | prename 's/(LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_\d{3})/$1.LUND/' LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_* |
</pre> | </pre> | ||
Revision as of 18:59, 14 August 2017
Since the LUND file is limited to 75000 particles (225000 lines), the gemc will have to be run in batch mode;
Creating a batch directory, with two subdirectories; 1)Phi_Parts, 2)submit.
1)Once the LUND file is broken into 178 parts, they can have the LUND extension added by:
prename 's/(Phi_Parts_\d{4})/$1.LUND/' Phi_Parts_*
Placing each of these files into its own directory, within a directory named Phi_Parts
find . -name "*.LUND" -exec sh -c 'mkdir "${1%.*}" ; mv "$1" "${1%.*}" ' _ {} \;
2)Creating the submit directory, and using a c++ program, creating the needed 178
#include <iomanip>
#include <sstream>
#include <iostream>
#include <fstream>
using namespace std;
void submit() {
for(int a=0;a<2;a++)
{
for(int b=0;b<10;b++)
{
for(int c=0;c<10;c++)
{
string filename="submit0";
stringstream hundreds;
hundreds << a;
stringstream tens;
tens << b;
stringstream ones;
ones << c;
string fullname="";
fullname=filename + hundreds.str() + tens.str() + ones.str();
// cout << fullname << "\n";
ofstream myfile;
myfile.open(fullname.c_str());
myfile << "#!/bin/sh\n";
myfile << "#PBS -l nodes=1\n";
myfile << "#PBS -A FIAC\n";
myfile << "#PBS -M vanwdani@isu.edu\n";
myfile << "#PBS -m abe\n";
myfile << "#\n";
myfile << "cd /home/lds/src/CLAS/GEMC\n";
myfile << "tcsh\n";
myfile << "source setup\n";
myfile << "cd /home/lds/src/GEANT/geant4.9.6/geant4.9.6-install/bin/geant4.sh\n";
myfile << "cd /home/vanwdani/src/GEANT4/geant4.9.6/Simulations/Research/Moller/batch/Phi_Parts/Phi_Parts_0";
myfile <<a<<b<<c<<"\n";
myfile << "gemc -USE_GUI=0 -Hall_Material=\"Vacuum\" -INPUT_GEN_FILE=\"LUND, Phi_Parts_0";
myfile <<a<<b<<c;
myfile << ".LUND\" -N=75000 eg12.gcard\n";
myfile << "~/src/CLAS/coatjava-1.0/bin/clas12-reconstruction -i eg12.ev -config DCHB::torus=1.0 ";
myfile << "-config DCHB::solenoid=0.0 -config DCTB::kalman=true -o eg12_rec.ev -s DCHB:DCTB:EC:FTOF:EB\n";
myfile << "~/src/CLAS/coatjava-1.0/bin/run-groovy Analysis.groovy eg12_rec.0.evio\n";
myfile.close();
}
}
}
}
This creates the submitXXXX file
#!/bin/sh #PBS -l nodes=1 #PBS -A FIAC #PBS -M vanwdani@isu.edu #PBS -m abe # cd /home/lds/src/CLAS/GEMC tcsh source setup cd /home/lds/src/GEANT/geant4.9.6/geant4.9.6-install/bin/geant4.sh cd /home/vanwdani/src/GEANT4/geant4.9.6/Simulations/Research/Moller/batch/Phi_Parts/Phi_Parts_0000 gemc -USE_GUI=0 -Hall_Material="Vacuum" -INPUT_GEN_FILE="LUND, Phi_Parts_0000.LUND" -N=75000 eg12.gcard ~/src/CLAS/coatjava-1.0/bin/clas12-reconstruction -i eg12.ev -config DCHB::torus=1.0 -config DCHB::solenoid=0.0 -config DCTB::kalman=true -o eg12_rec.ev -s DCHB:DCTB:EC:FTOF:EB ~/src/CLAS/coatjava-1.0/bin/run-groovy Analysis.groovy eg12_rec.0.evio
Creating a file named lds-submit
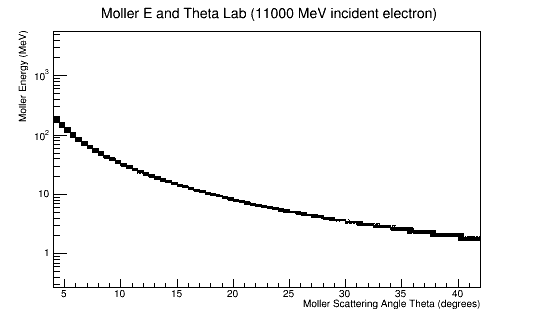
11GeV incident electron
Simulating with GEMC
For an incident electron at 11GeV, the Moller electron in the range of 5 to 40 degrees for the scattering angle Theta, has energies lower than 200MeV.
~/src/CLAS/GEMC/experiments/eg12/MolrBckGrd/DV/Isotropic_study/LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut> wc -l LUND_Spread_LH2_IsotropicPhi.LUND 315000 LUND_Spread_LH2_IsotropicPhi.LUND
Spliting the LUND file into 105 parts of 1000 events, or 3000 lines
split -d -l 3000 -a 3 LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut.LUND LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_
Naming these files LUND files
prename 's/(LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_\d{3})/$1.LUND/' LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut_*
Placing each of these files into its own directory, within a directory named LH2_0Sol_0Tor_11GeV_IsotropicPhi_ShieldOut
find . -name "*.LUND" -exec sh -c 'mkdir "${1%.*}" ; mv "$1" "${1%.*}" ' _ {} \;
Defining the gcards
<gcard>
<!-- minumal detectors for HB tracking -->
<detector name="../../../../../../clas12/fc/forwardCarriage" factory="TEXT" variation="original"/>
<detector name="../../../../../../clas12/dc/dc" factory="TEXT" variation="ccdb"/>
<detector name="../../../../../../clas12/ftof/ftof" factory="TEXT" variation="java"/>
<option name="SCALE_FIELD" value="clas12-torus-big, -1"/>
<option name="OUTPUT" value="evio, Molr.evio"/>
<!-- Solenoid apparatus and field -->
<!--detector name="../../../../../../clas12/magnets/solenoid" factory="TEXT" variation="original"/-->
<!--option name="HALL_FIELD" value="clas12-solenoid"/-->
<!-- other CLAS detectors -->
<detector name="../../../../../../clas12/ec/ec" factory="TEXT" variation="original"/>
<detector name="../../../../../../clas12/ctof/ctof" factory="TEXT" variation="original"/>
<detector name="../../../../../../clas12/htcc/htcc" factory="TEXT" variation="original"/>
<detector name="../../../../../../clas12/pcal/pcal" factory="TEXT" variation="javageom"/>
<!--detector name="../../../../../../clas12/micromegas/micromegas" factory="TEXT" variation="original"/-->
<!-- Beamline material -->
<!--detector name="../../../../../../clas12/beamline/beamline" factory="TEXT" variation="ft"/-->
</gcard>
Using a file to store the needed commands
/src/CLAS/GEMC/source/gemc -USE_GUI=0 -INPUT_GEN_FILE="LUND,MolTest.LUND" -N=100 eg12.gcard Solenoid off ~/src/CLAS/coatjava-2.4/bin/clas12-reconstruction -i ../files/MolrBckGrd/Molr.evio -config GEOM::new=true -config MAG::torus=1.0 -config MAG::solenoid=0.0 -o ../files/MolrBckGrd/Molr_rec.evio -s DCHB:DCTB:EC:FTOF:EB -config DATA::mc=true Solenoid ON ~/src/CLAS/coatjava-2.4/bin/clas12-reconstruction -i ../files/MolrBckGrd/Molr.evio -config GEOM::new=true -config MAG::torus=-1.0 -config MAG::solenoid=1.0 -o ../files/MolrBckGrd/Molr_rec.evio -s DCHB:DCTB:EC:FTOF:EB -config DATA::mc=true -config DCTB::useRaster=true ~/src/CLAS/coatjava-2.4/bin/evio-dump -i ../files/MolrBckGrd/Molr_rec.0.evio
Analyzing Results
Using cat to combine the reconstructed particles found using Analysis.groovy, we can output the particle number from the LUND file, Total Energy, scattering angles Theta, and Phi for the generated and reconstructed particles respectively.
For 0T:
571-1 0.12103204139108564 5.232292493596922 9.99999999997547 0.2397734 64.37257 163.2439 23025-3 0.02376743729734356 11.825408412314742 9.999999999978474 0.29301766 89.180336 -164.40204
For 5T:
24726-1 0.08576327895104396 6.223977074710134 9.99999999997547 0.17311224 20.298353 39.036583 19970-3 0.026540097007028158 11.193535798440614 9.999999999969427 0.15106007 49.16122 14.2969885
Converting the particle number per run into the particle number via the LUND file:
Multiplying the line number by 3 accounts for the 3 lines per particle in the LUND file. From this LUND particle we find the weight associated with each particle (Specific Energy and angle Theta).
To find the number of particles we can use an awk command:
awk 'NR == 1711 { print $6 }' LUND_Spread.LUND
| Particle number | Run number | LUND Moller electron number | Overall LUND particle number | Corrected LUND header line number | Number of Particles |
|---|---|---|---|---|---|
| 571 | 1 | 571 | 1713 | 1711 | 1097.227223687062 |
| 23025 | 3 | 69075 | 207225 | 207223 | 25951.273801947842 |
| 24726 | 1 | 24726 | 74178 | 74176 | 2579.181846387747 |
| 19970 | 3 | 59910 | 179730 | 179728 | 14255.222465310777 |
Cross Section
Using the number of expected Moller electrons and the fact that the reaction cross section can be found using the Luminosity we find: Integrating the differential cross-section over the solid angle:
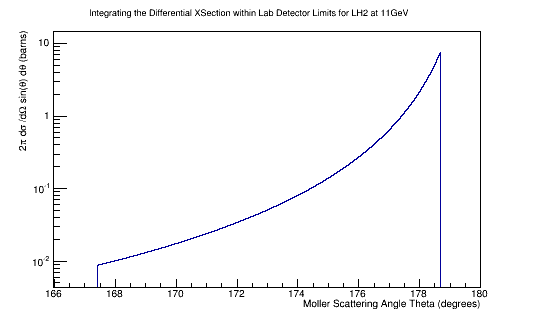
IntegralDiffXSect->Integral()
4.97493824519086629e+04 barns
Multiplying by the Luminosity, we find:
Since this is just a ratio of detected particles to total particles, this gives the cross section as a relative probablity of a scattering, or reaction, to occur.
For 0T with 11GeV incident electrons:
For 5T with 11GeV incident electrons:
5.5GeV incident electron
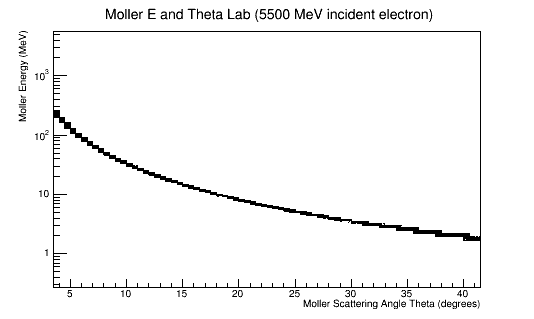
For 0T:
16544-1 0.10778422619732184 5.519947956962607 9.999999999974188 0.19995739 25.982777 96.17941
For 5T:
354-1 0.09640233076149667 5.8423283009263045 9.999999999976643 0.09051078 49.163692 -85.56053 5526-4 0.03948624565299794 9.164813676343726 9.999999999971514 0.12387912 52.14526 50.051956 21892-4 0.02741437437384611 11.000629446532491 9.99999999997668 0.26457193 81.77688 179.14293
| Particle number | Run number | LUND Moller electron number | Overall LUND particle number | Corrected LUND header line number | Number of Particles |
|---|---|---|---|---|---|
| 16544 | 1 | 16544 | 49632 | 49630 | 814.898350440404 |
| 354 | 1 | 354 | 1062 | 1060 | 556.656736645754 |
| 5526 | 4 | 22104 | 66312 | 66310 | 937.328333457728 |
| 21892 | 4 | 87568 | 262704 | 262702 | 8343.893471134810 |
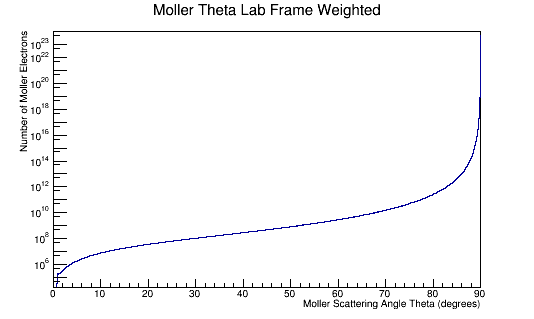
MolThetaLabweighted->Integral()
This gives about 4.96e+23 incident electrons
For 0T with 5.5GeV incident electrons:
For 5T with 5.5GeV incident electrons: