October 22, 2008
Unpolarized Measurents:
A 2x4x8" aluminum brick was placed at the HRRL export in order to destroy the photon polarization. Rate measurements were made with the descriminator and HV settings used the past week. Then the constant fraction descriminator was replaced with a threshold descriminator on two detectors but not the reference detector.
ADC =0 => Reference Detector D
ADC = 1 => NaI
ADC=2 => Detector C
ADC = 3 => Detector A
channel # * 0.1675 ns/chan => ns
Default HV and CF descriminator
Matched HV and Threshold Descriminator
2 Runs were taken with the following settings
100 ms Threshold descrimnaotr
=
=
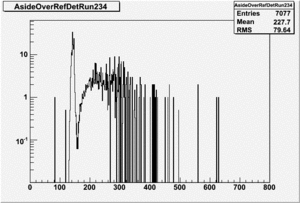
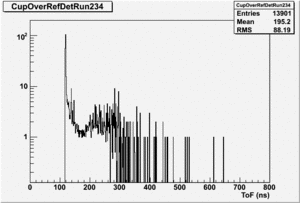
Run 168 ( 234.lst)
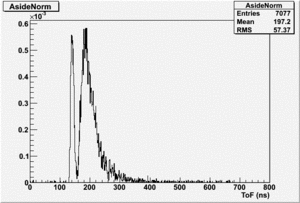
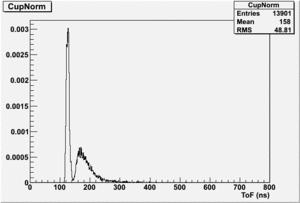
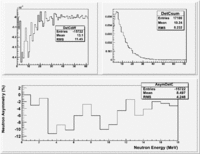
If I normalize using the Reference detector I see
If I normalize using the NaI detector I see:
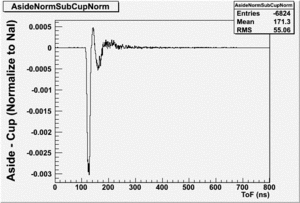
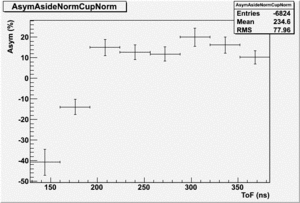
The Difference and Asymmetry measurements.
The above is just an example of how to analyze the data. Students should take my bad example and improve upon it.
The program I used to generate the above pictures from a ROOT file is given below.
I used the "PhotoFis->MakeClass()" command within root to generate the skeleton program which has 2 files called Media:PhotoFis.h.txt and Media:PhotoFis.C.txt.
You define your histogram variables and the file you are opening to analyze in the file Media:PhotoFis.h.txt.
see the lines which look like
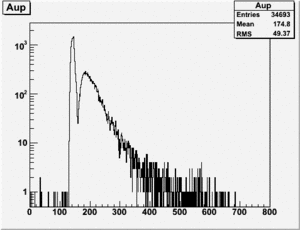
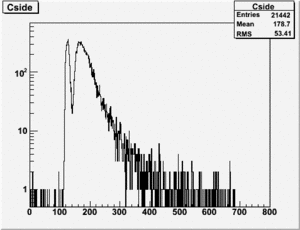
TH1F *NaiInt229, *RefDetRun229, *Aup, *Cside, *Nai229, *Unity; TH1F *NaiInt234, *RefDetRun234, *Aside, *Cup, *Nai234; TH1F *AsideOverRefDetRun234, *CupOverRefDet234, , *AupOverRefDet229, *CsideOverRefDet229; TH1F *AsideNormSubCupNorm, *AsideNormAddCupNorm, *AsymAsideNormCupNorm;
TFile *f = (TFile*)gROOT->GetListOfFiles()->FindObject("DetCup_10_10_08_00234.root");
if (!f) {
f = new TFile("DetCup_10_10_08_00234.root");
You create your histograms, loop over the ntuple entries , and perform histogram operations in the file Media:PhotoFis.C.txt
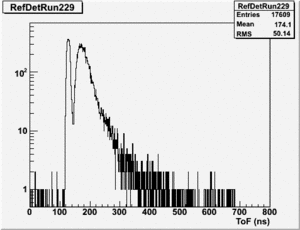
Run 164 (229.lst)
The root commands
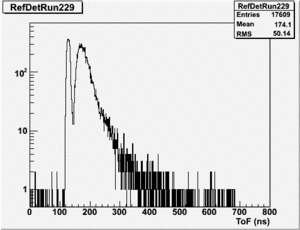
root [24] TH1F *RefDetRun229=new TH1F("RefDetRun229","RefDetRun229",800,0,800);
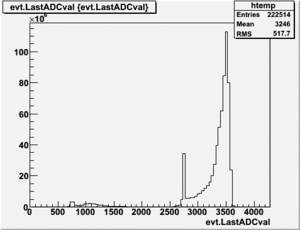
root [25] PhotoFis->Draw("evt.ADCval*0.1675 >> RefDetRun229","evt.ADCnum==0 && evt.LastADCval>0");
Will create histogram with 800 channels and 800 bins so we have 1 bin per channel (ns).
To normalize the data you need to integrate the NaI histogram. To do this a skeleton program is creates using ROOT which will loop through the ROOT ntuple using the command
PhotoFis->MakeClass();
You will see a file called PhotoFis.C after you execute the above command within the ROOT interpreter window.
Asym
Now do Asym plots using the same detector normalize by the Reference detector in one case and the NaI detector in another.
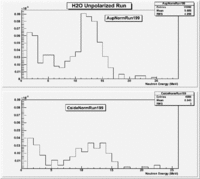
H20 runs
Run 140 (199.lst)
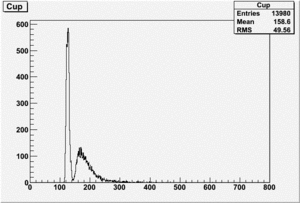
Det A on top
uses file 199.lst
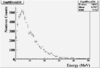
The spectrum below shows the Time of Flight spectrum for Detectors "A" and "C". The bottom figures represent the energy calculated from the time of flight when a neutron is assumed as the detected particle. Events beyond 15 MeV are from the photon peak (phtotons are misidentified as neutrons). In order to determine the time of flight the average ToF for a photon (140 ns) was used as the start time. Photon events above this average will appear at energies beyond 15 MeV and photon events below this average will have negative times and appear as Underflows in histogram AupNErun199. There are 7640 events in Histogram Auprun199 above 146 ns correspond to the (15050-7410=7640) events shown in AupNErun199 histogram.
Neutrons from the target should have energies below 14 MeV. The spectra below were normalized by the number of NaI events observed during the run.
Run 136 (194.lst)
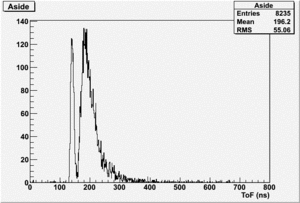
Det A on side
uses file 194.lst
ROOT Commands
1.)How to save histograms from several different files to a single file
root [0] TFile *f1=new TFile("Run194Histos.root")
root [1] TH1F *AsideNormRun194_H=((TH1F*)f1->Get("AsideNormRun194"));
root [2] TFile* file = TFile::Open("UnpolResults.root","RECREATE");
root [3] file->cd();
root [4] AsideNormRun194_H->Write();
root [5] TFile *f2=new TFile("Run199Histos.root")
root [6] TH1F *AupNormRun199_H=((TH1F*)f2->Get("AupNormRun199"));
root [7] file->cd();
root [8] AupNormRun199_H->Write();
2.) How to manipulate histograms in the saved file
TFile *f1=new TFile("UnpolResults.root");
f1->cd();
TH1F *AupNormRun199_H=((TH1F*)f1->Get("AupNormRun199"));
AupNormRun199_H->Draw();
TH1F *AsideNormRun194_H=((TH1F*)f1->Get("AsideNormRun194"));
AsideNormRun194_H->Draw();
TH1F *AsideNormRun194_H=((TH1F*)f1->Get("AsideNormRun194"));
AupNormRun199_H->Sumw2();
AsideNormRun194_H->Sumw2();
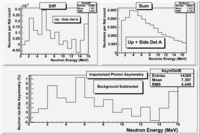
TH1F H2OdiffDetA= (*AupNormRun199_H - *AsideNormRun194_H);
H2OdiffDetA->Draw();
TH1F H2OsumDetA= (*AupNormRun199_H + *AsideNormRun194_H);
H2OsumDetA->Draw();
Background subtracted Det A
TFile *f1=new TFile("Run194Histos.root"); // H2O Aside
TFile *f2=new TFile("Run199Histos.root"); // H2O Aup
TFile *f3=new TFile("Run229Histos.root"); // D2O Aup
TFile *f4=new TFile("Run234Histos.root"); // D2O A side
f1->cd();
TH1F *AsideNormRun194=((TH1F*)f1->Get("AsideNormRun194"));
f2->cd();
TH1F *AupNormRun199=((TH1F*)f2->Get("AupNormRun199"));
f3->cd();
TH1F *AupNormRun229=((TH1F*)f3->Get("AupNormRun229"));
f4->cd();
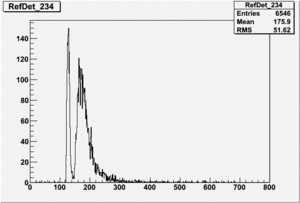
TH1F *AsideNormRun234=((TH1F*)f4->Get("AsideNormRun234"));
TFile* file = TFile::Open("UnpolResultsDetA.root","RECREATE");
file->cd();
AsideNormRun194->Write();
AupNormRun199->Write();
AupNormRun229->Write();
AsideNormRun234->Write();
AsideNormRun194->Sumw2();
AupNormRun199->Sumw2();
AupNormRun229->Sumw2();
AsideNormRun234->Sumw2();
TH1F DetAdiffH20= ( *AupNormRun199 - *AsideNormRun194);
DetAdiffH20->SetName("DetAdiffH20");
TH1F DetAup_D2OsubH20= ( *AupNormRun229 - *AupNormRun199);
DetAup_D2OsubH20->SetName("DetAup_D2OsubH20");
TH1F DetAside_D2OsubH20= ( *AsideNormRun234 - *AsideNormRun194);
DetAside_D2OsubH20->SetName("DetAside_D2OsubH20");
TH1F DetAdiff= (DetAup_D2OsubH20 - DetAside_D2OsubH20);
DetAdiff->SetName("DetAdiff");
DetAdiffH20->Write();
TH1F DetAsum= (DetAup_D2OsubH20 + DetAside_D2OsubH20);
DetAsum->SetName("DetAsum");
DetAsumH20->Write();
DetAup_D2OsubH20->Write(); DetAside_D2OsubH20->Write(); DetAdiff->Write();
TH1F *AsymDetA=(TH1F*)DetAdiff->Clone("AsymDetA");
AsymDetB->Scale(25000); // 100 * 0.004 from the DetA sum histogram
Solid Angle Asymmetry
The "Up/Side" asymmetry is measured by positioning one detector a distance vertically above the beam and another detector horizontally displaced a distance away from the beam. The solid angle subtended by the PMTs used to detect neutrons is calculated in terms of the surface Area of the PMT face divided by the PMT distance from the target squared.
A geometrical asymmetry is induced if you position the PMTs at difference distances from the target. Assuming that both detectors have the same surface area (A), the "Up" rate - "Side" rate asymmetry may be written as:
- Asymm
For the above measurements the "Up" detector was a distance of 244.775 cm and the "Side" detector was a distance of 232.131 cm from the target. The detectors were suppose to be 231.775 cm from the target when running with polarized photons.
The solid angle asymmetry is
- Asym
T2E (Convert time to Neutron Energy)
Float_t T2E(Int_t UpDown, Int_t TDCval)
{
Float_t Ekin,Beta,Gamma,Eneutron,Tgamma,SpeedLightcm;
Float_t Dist,polDist,UnPolTopDist,UnPolSideDist;
polDist = 231.775; //cm
UnPolTopDist = 244.775; //cm
UnPolSideDist = 232.131; //cm
SpeedLightcm=30; // cm/ns
Tgamma=146/0.1675;
Eneutron=939.5656;
if(UpDown==0)
Dist=polDist;
if(UpDown==1)
{
Dist=UnPolTopDist;
Tgamma=146/0.1675;
}
if(UpDown==2)
{
Dist=UnPolSideDist;
Tgamma=130/0.1675;
}
// printf("TDCval=%g - Tgamma = %g\n",TDCval,Tgamma);
Beta=(TDCval-Tgamma)*0.1676;
if(Beta<0)
return(-1.0);
Beta=1/(1+Beta*SpeedLightcm/Dist);
// printf("Beta=%g\n",Beta);
Gamma=1/(1-Beta*Beta);
if(Gamma<0)
return(-2.0);
Gamma=sqrt(Gamma);
Ekin=(Gamma-1)*Eneutron;
return(Ekin);
}
Results Histogram Table
|
Desc |
Det A up | Det A side | Det C up | Det C side | ||||
| D2O | H2O | D2O | H2O | D2O | H2O | D2O | H2O | |
| ToF | 
|

|

|
|
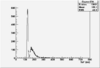
|
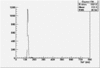
|
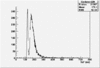
|
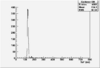
|
| Energy | 
|

|

|

|
|

|

|

|
| Energy Norm by NaI | 
|

|

|

|

|

|

|

|
| Backgrnd Subtraction | 100px | 100px | 
|

| ||||
| Backgrnd Subtraction | 100px | 100px | 
|

| ||||
| Asymmetry | 100px | 
| ||||||
Error calculation for 4 MeV neutrons
Cup(D2O) = 480 counts
Cup(H2O) = 9 counts
Cside (D2O) = 1300 Counts
Cside (H2O) = 4 Counts
Asym
= 0.1 %
Still need to double check the above. This is the best possible error when assuming it should be \frac{\sigma}{N} but I don't know what is yet.