Difference between revisions of "PePPO 03132012"
Jump to navigation
Jump to search
| (28 intermediate revisions by 2 users not shown) | |||
| Line 67: | Line 67: | ||
{| border="1" |cellpadding="20" cellspacing="0 | {| border="1" |cellpadding="20" cellspacing="0 | ||
|- | |- | ||
| − | | Run#|| run mode|| Run length (sec)||Beam||Target ||Trig Logic|| VME discriminator Threshold (mV)|| HV1 (setting/V) || HV2 (setting/V) || | + | | Run#|| run mode|| Run length (sec)||Beam||Target ||Trig Logic|| VME discriminator Threshold (mV)|| HV1 (setting/V) || HV2 (setting/V) || Deadtime || |
|- | |- | ||
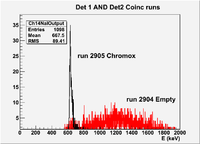
| − | | | + | | 2902|| sample || 0 || e<math>^-</math> || Chromox || (Det1 OR Det2) || 50 || 2.4/960 || 3/1200 || 93.1818% <math>\pm</math> 10.3874% || |
|- | |- | ||
| − | | | + | | 2903|| sample || 203 ||e<math>^-</math> || Empty || (Det1 OR Det2) || 50 || 2.4/960 || 3/1200 || 84.9788% <math>\pm</math>0.106622% || |
|- | |- | ||
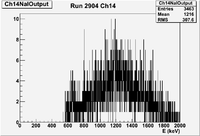
| − | | | + | | 2904|| sample || 452 ||e<math>^-</math> || Empty || Det1 AND Det2 || 50 || 2.4/960 || 3/1200 || 12.2563% <math>\pm</math> 2.32772% || |
|- | |- | ||
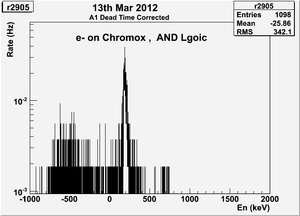
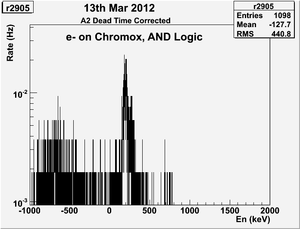
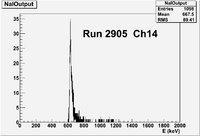
| − | | | + | | 2905|| sample || 556 || e<math>^-</math> || Chromox || Det1 AND Det2 || 50 || 2.4/960 || 3/1200 || 3.16901% <math>\pm</math> 4.2301% || [[File:peppo-032302012-r2905-A1.png | 300 px]] [[File:peppo-032302012-r2905-A2.png | 300 px]] |
|- | |- | ||
| − | | | + | | 2908|| sample || 150 || off || out || (Det1 or Det2) || 50 || 2.4/960 || 3/1200 || 74.0667% <math>\pm</math> 0.157206% || |
|- | |- | ||
| − | | | + | | 2909|| sample || 1118 || off || out || (Det1 AND Det2) || 50 || 2.4/960 || 3/1200 || 3.57143% <math>\pm</math>26.9725% || |
|- | |- | ||
|} | |} | ||
| + | |||
| + | |||
| + | = data analysis = | ||
| + | |||
| + | ==Negative Energy ? == | ||
| + | |||
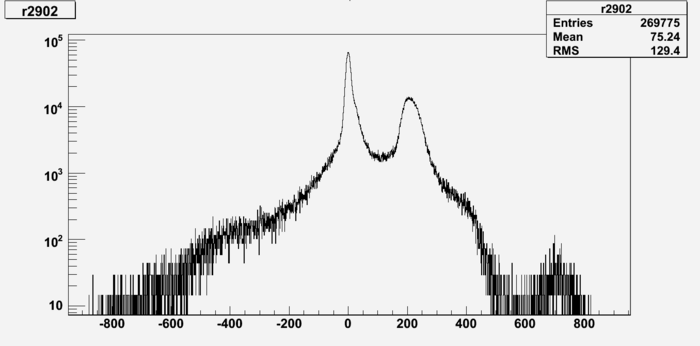
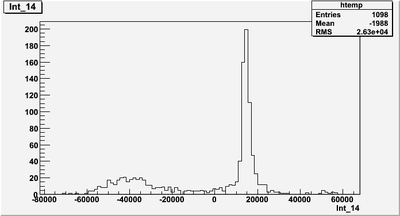
| + | This run 2902, Int_Ch14 histogram. Why there is negative? | ||
| + | |||
| + | [[File:peppo-032302012-r2902-int14-negative.png | 700 px]] | ||
| + | |||
| + | == Pulses == | ||
| + | |||
| + | === run 2905 === | ||
| + | |||
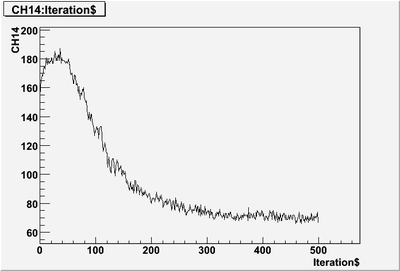
| + | 1st 100 puleses | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-1to100.png | 400 px]] | ||
| + | |||
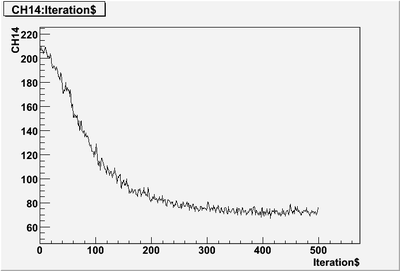
| + | pulse that only partially read-in: | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-1-pulse-cut-off.png | 400 px]] | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-9th-pulse-cut-off.png | 400 px]] | ||
| + | |||
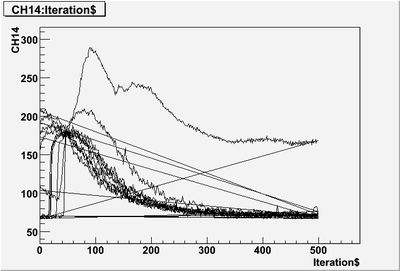
| + | first 10 pulses, some pulses that only partially read-in: | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-10-pulse-cut-off.png | 400 px]] | ||
| + | |||
| + | |||
| + | looks like some pulses are only partially read-in. We need to delay signal more? | ||
| + | |||
| + | One possible cut may to determine if the signal is increasing within the first 100 - 300 $Iterations. This would be like creating a leading edge discriminator in software. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
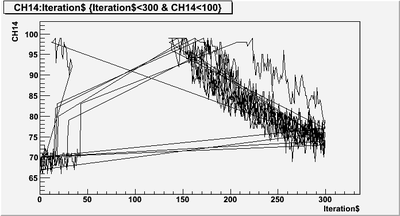
| + | T->Draw("CH14:Iteration$","Iteration$<300 & CH14<100","L",10,1) | ||
| + | |||
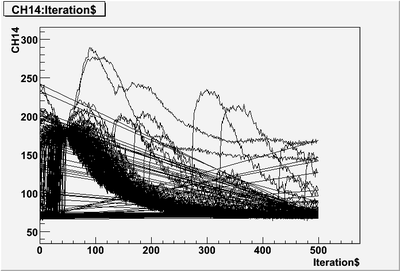
| + | looks like it just won't draw. I need to know the to way exclude that event does not satisfy "Iteration$<300 & CH14<100" requirement. | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-selection-try-1.png | 400 px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
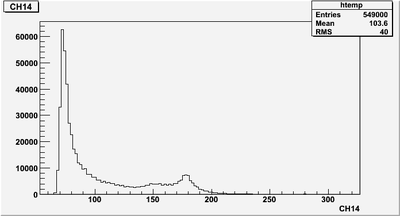
| + | Why CH14 histogram is all positive, but int_14 histogram has negatives? | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-CH14-Hist.png | 400 px]] | ||
| + | |||
| + | [[File:PePPO-03132012-pulses-2905-int_14-Hist.png | 400 px]] | ||
| + | |||
| + | ==TF Analysis== | ||
| + | |||
| + | |||
| + | Using the following ROOT programs | ||
| + | |||
| + | [[Media:03212012_T.C.txt]] | ||
| + | |||
| + | [[Media:03212012_T.h.txt]] | ||
| + | |||
| + | |||
| + | |||
| + | {| border="1" |cellpadding="20" cellspacing="0 | ||
| + | |- | ||
| + | | Run#|| run mode|| Run length (sec)||Beam||Target ||Trig Logic|| VME discriminator Threshold (mV)|| HV1 (setting/V) || HV2 (setting/V) || Deadtime || | ||
| + | |- | ||
| + | | 2902|| sample || 0 || e<math>^-</math> || Chromox || (Det1 OR Det2) || 50 || 2.4/960 || 3/1200 || 93.1818% <math>\pm</math> 10.3874% || | ||
| + | |- | ||
| + | | 2903|| sample || 203 ||e<math>^-</math> || Empty || (Det1 OR Det2) || 50 || 2.4/960 || 3/1200 || 84.9788% <math>\pm</math>0.106622% || | ||
| + | |- | ||
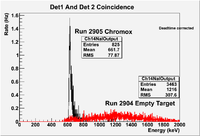
| + | | 2904|| sample || 452 ||e<math>^-</math> || Empty || Det1 AND Det2 || 50 || 2.4/960 || 3/1200 || 12.2563% <math>\pm</math> 2.32772% || [[File:03212012_Ch14r2904.png| 200 px]] | ||
| + | |- | ||
| + | | 2905|| sample || 556 || e<math>^-</math> || Chromox || Det1 AND Det2 || 50 || 2.4/960 || 3/1200 || 3.16901% <math>\pm</math> 4.2301% || [[File:03212012_Ch14r2905.png| 200 px]] | ||
| + | |- | ||
| + | | 2908|| sample || 150 || off || out || (Det1 or Det2) || 50 || 2.4/960 || 3/1200 || 74.0667% <math>\pm</math> 0.157206% || [[File:03212012_Ch14r2908.png| 200 px]] | ||
| + | |- | ||
| + | | 2909|| sample || 1118 || off || out || (Det1 AND Det2) || 50 || 2.4/960 || 3/1200 || 3.57143% <math>\pm</math>26.9725% || | ||
| + | |- | ||
| + | |||
| + | |} | ||
| + | |||
| + | Run 2904 and 2905 coincidence overlay (NOT Deadtime corrected) | ||
| + | |||
| + | [[File:03212012_Coinc2904-5.png | 200 px]] | ||
| + | |||
| + | Normalizing using run time and correcting for DAQ deadtime: | ||
| + | |||
| + | [[File:03212012_NormCoinc2904-5.png | 200 px]] | ||
Latest revision as of 06:35, 22 March 2012
Accelerator Configuration
- Accelerator Settings
| beam energy | 6.8 MeV |
| Beam type: | Electron |
| I_average: | 0.3 nA |
| I_peak: | 0.05 A 0.006 duty factor |
| pulse width: | 10 s (it seems on 200 s is possible?) |
| Rate: | 60 Hz |
| PEPPO Spectrometer: | 151.3 A |
Set HV to Detectors
| Signal channel | HV dac | HV (V) | |
| Detectors #1 | A1 | 2.4 | 960 |
| Detectors #2 | A2 | 3.0 | 1200 |
Scope Pictures
CH1: Beam Sync CH2: A1 CH3: A2
| Target | Pictures |
| No Target | 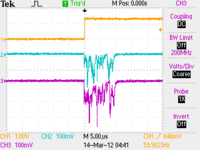 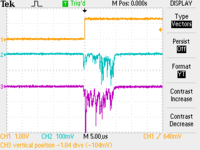
|
| Chromox Target | 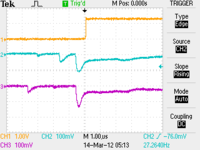 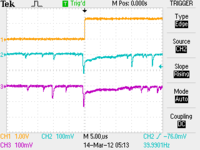
|
Annihilation Counter Runs
- Signal Definitions for Trigger logic
- SYNC:=60 Hz sync pulse 200 micro seconds long
- Det1:= Annihilation Counter #1 (HV1)
- Det2:=Annihilation Counter #2 (HV2)
data analysis
Negative Energy ?
This run 2902, Int_Ch14 histogram. Why there is negative?
Pulses
run 2905
1st 100 puleses
pulse that only partially read-in:
first 10 pulses, some pulses that only partially read-in:
looks like some pulses are only partially read-in. We need to delay signal more?
One possible cut may to determine if the signal is increasing within the first 100 - 300 $Iterations. This would be like creating a leading edge discriminator in software.
T->Draw("CH14:Iteration$","Iteration$<300 & CH14<100","L",10,1)
looks like it just won't draw. I need to know the to way exclude that event does not satisfy "Iteration$<300 & CH14<100" requirement.
Why CH14 histogram is all positive, but int_14 histogram has negatives?
TF Analysis
Using the following ROOT programs
Run 2904 and 2905 coincidence overlay (NOT Deadtime corrected)
Normalizing using run time and correcting for DAQ deadtime: